Abstract
Adaptive responses during phosphate (Pi) starvation are regulated by complex molecular mechanisms in plants. Transcription factors are believed to be the key determinants of Pi starvation responses. We have recently identified the plant-specific WRKY75 transcription factor as an important component of the Pi stress responses. WRKY75 is a positive regulator of several phosphate starvation induced (PSI) genes including phosphatases, Mt4/TPS1-like genes and high affinity Pi transporters. It also acts as a negative regulator of some components of root development, independent of Pi stress response. WRKY75 has considerable effect on anthocyanin accumulation, Pi uptake and Pi content in the plant. Here we present a hypothetical model of transcriptional regulation during phosphate starvation induced processes in plants which help in the maintenance of Pi homeostasis.
Key Words: Arabidopsis, transcription factors, phosphate stress, root architecture, WRKY75
Biological Function of the WRKY75 Transcription Factor
Phosphorus (P) is a vital nutrient required for numerous metabolic and developmental processes in plants. However, its availability in most soils is limited as it is fixed in mineral or organic forms that are unavailable to plants.1 Plants respond to the lack of readily available Pi through adaptive modifications such as altered root architecture,2 elevated phosphatase activity,3 secretion of organic acids1 and increased expression of Pi transporters.4 Together, these adaptive mechanisms help plants improve their ability to mobilize, acquire and utilize Pi efficiently.5 However, the molecular determinants regulating these adaptive mechanisms are yet to be completely revealed. Until recently PHR1, a MYB transcription factor that controls a small subset of Pi starvation responses,6 was the only known regulator of Pi starvation stress responses in Arabidopsis.
We used data from our earlier microarray study of PSI genes7 to identify several putative Pi responsive transcription factors (TFs). WRKY75, a plant-specific transcription factor was one of the candidate TFs that was strongly induced during Pi deprivation and localized to the nucleus irrespective of the Pi status of the plant. To decipher the in planta role of WRKY75 in Pi stress responses, we suppressed its expression through RNAi silencing. The mutant plants demonstrated early accumulation of anthocyanin during Pi deprivation, suggesting an impaired Pi stress response mechanism. Further analysis revealed that the suppression of WRKY75 led to a variable, but significant, decrease in the expression of several key PSI genes. These genes included phosphatases AtPS2-1 and AtPS2-2, high affinity phosphate transporters Pht1;1 and Pht1;4,8 as well as At4 and IPS1 which are presumed to be involved in Pi translocation and signaling during Pi stress.9,10 This resulted in decreased uptake of Pi leading to reduced Pi content in the plant. The data indicated that WRKY75 is a positive regulator of several genes that are vital for mobilizing, acquiring and translocating Pi as well as signaling during Pi deprivation. WRKY TFs are known to regulate their target genes by specifically binding to TTGACC/T (W-box) elements on the promoters of these genes.11,12 In silico analysis revealed that a large number of PSI genes had W-Box elements on their promoters. The expression level of PSI genes in WRKY75 RNAi mutants was strongly correlated with the number and type of W-Box on the promoters of the PSI genes. Interestingly, the presence of W-boxes with the TTGACC sequence appeared to have a greater effect than those with the TTGACT sequence, suggesting a fine tuning of regulation of PSI genes by WRKY75. The suppression of WRKY75 significantly increased the lateral root length, density and the numbers of root hairs, thereby helping the mutant plants acquire more Pi over a longer period. The regulation of root development by WRKY75 occurred irrespective of the Pi status in the RNAi mutant plant. This suggests that WRKY75 also functions as a negative regulator of root development independent of Pi starvation status. Based on these results, we postulate that WRKY75 functions as a constitutive negative regulator of root development and thus influences Pi uptake. On the other hand, during Pi starvation, it functions as a positive regulator of PSI genes. Taken together, these data indicates that WRKY75 has a major role in the maintenance of Pi homeostasis within the plant.
Transcriptional Regulation of Pi Starvation Responses: A Hypothetical Model
The regulation of Pi stress responses by the PHR1 transcription factor has been well characterized in Arabidopsis.6,13 Our studies showed that WRKY75 regulates several aspects of Pi starvation independent of PHR1.14 Considerable progress has also been made in characterizing ZAT6 and MYB62, two other Pi stress responsive transcription factors that we identified recently (unpublished data, BND and KGR). It is now clear that the regulation of PSI genes and root architecture is fundamental for the maintenance of Pi homeostasis. Therefore, based on the functions of the currently known molecular determinants, we propose a hypothetical model for the transcriptional regulation of root architecture and PSI genes during Pi deprivation (Fig. 1). In this model, Pi deprivation induces the expression of several transcription factors. PHR1 is regulated post-translationally through sumoylation by SIZ1, a SUMO E3 ligase.15 A previous report has shown that PHR1 regulates the expression of PSI genes directly as well as through the recently discovered microRNA, miR399.13 On the other hand, our data suggests that ZAT6 and MYB62 regulate root architecture, possibly through phytohormones, in response to Pi starvation. MYB62 also contains two SUMO domains suggesting that it is a potential sumoylation candidate for SIZ1. Interestingly, WRKY75 independently regulates both root architecture as well as PSI gene expression. López-Bucio et al.,2 have provided a concise review of the role played by the phytohormones auxin, cytokinin and ethylene in the regulation of root architecture during Pi starvation stress. The role of sugars in regulating Pi homeostasis and root architecture has also been clearly demonstrated.16,17 We therefore propose that ZAT6, MYB62 and WRKY75 regulate root architecture through the modulation of signaling compounds such as phytohormones and sugars. Abel et al.,18 have proposed the presence of a hypothetical intracellular phosphate sensing mechanism that regulates phosphate starvation responses. In support of this notion, we found that ZAT6 influences PSI gene expression by modulating the intracellular Pi content through the regulation of root architecture. Therefore, we speculate that the maintenance of Pi homeostasis through transcriptional regulation is a complex bi-directional process involving the modulation of root architecture and PSI gene expression in tandem.
Figure 1.
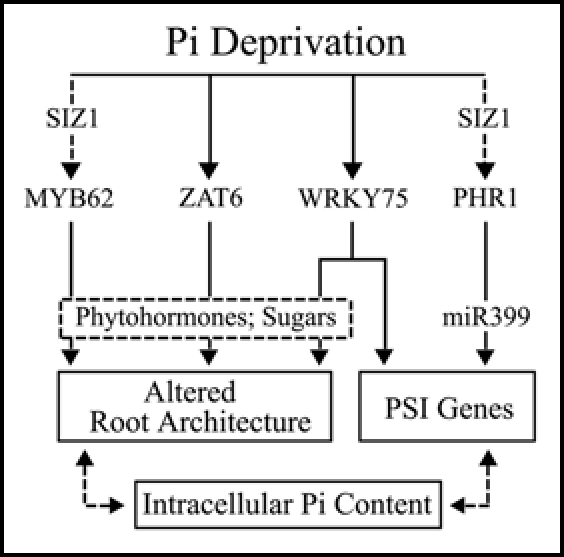
Schematic representation of a hypothetical model for the transcription regulation of Pi homeostasis. This model proposes the likely position of several Pi responsive transcription factors within the overall regulatory setup controlling Pi homeostasis in Arabidopsis.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4418
References
- 1.Marschner H. Mineral nutrition of higher plants. London: Academic Press; 1995. [Google Scholar]
- 2.López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role nutrient availability in regulating root architecture. Curr Opin Plant Biol. 2003;6:280–287. doi: 10.1016/s1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- 3.Lipton DS, Lanchar RW, Blevins DG. Citrate, malate and succinate concentration in exudates from P-sufficient and P-stressed Medicago sativa L. seedlings. Plant Physiol. 1987;85:315–317. doi: 10.1104/pp.85.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghothama KG, Karthikeyan AS. Phosphate acquisition. Plant Soil. 2005;274:37–49. [Google Scholar]
- 5.Rausch C, Bucher M. Molecular mechanisms of phosphate transport in plants. Planta. 2002;216:23–27. doi: 10.1007/s00425-002-0921-3. [DOI] [PubMed] [Google Scholar]
- 6.Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signalling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrerra-Estrella L, Nussaume L, Thibaud MC. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muchhal US, Pardo JM, Raghothama KG. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin H, Shin HS, Chen R, Harrison MJ. Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J. 2006;45:712–726. doi: 10.1111/j.1365-313X.2005.02629.x. [DOI] [PubMed] [Google Scholar]
- 10.Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, De La Peña A, Leyva A, Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000;24:1–11. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- 11.Ulker B, Somssich IE. WRKY transcription factors: From DNA binding towards biological function. Curr Opin Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Kim KC, Fan B, Chen Z. Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 2006;142:1180–1192. doi: 10.1104/pp.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bari R, Pant BD, Stitt M, Scheible WR. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun D, Hasegawa PM. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karthikeyan AS, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta. 2006;225:907–918. doi: 10.1007/s00425-006-0408-8. [DOI] [PubMed] [Google Scholar]
- 17.Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG. Differential effects of sucrose and auxin on localized Pi-deficiency induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol. 2007 doi: 10.1104/pp.106.092130. online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abel S, Ticconi AC, Delatorre CA. Phosphate sensing in higher plants. Physiol Plant. 2002;115:1–8. doi: 10.1034/j.1399-3054.2002.1150101.x. [DOI] [PubMed] [Google Scholar]


