Abstract
Antisense oligodeoxynucleotide (ODN) inhibition emerges as an effective means for probing gene function in plant cells. Employing this method we have established the importance of the SUSIBA2 transcription factor for regulation of starch synthesis in barley endosperm, and arrived at a model for the role of the SUSIBAs in sugar signaling and source-sink commutation during cereal endosperm development. In this addendum we provide additional data demonstrating the suitability of the antisense ODN technology in studies on starch branching enzyme activities in barley leaves. We also comment on the mechanism for ODN uptake in plant cells.
Key words: antisense ODN, barley, SBE, SUSIBA2
Antisense ODNs are short (12–25 nt-long) stretches of single-stranded ODNs that hybridize to the cognate mRNA in a sequence-specific manner, thereby inhibiting gene expression. They are naturally occurring in both prokaryotes and eukaryotes where they partake in gene regulation and defense against viral infection.1,2 The mechanisms for antisense ODN inhibition are not fully understood but it is generally considered that the ODN either sterically interferes with translation or promotes transcript degradation by RNase H activation.3,4
The earliest indication of the usefulness of antisense ODN technology for the purposes of molecular biology and medical therapy was the demonstration in 1978 that synthetic ODNs complementary to Raos sarcoma virus could inhibit virus replication in tissue cultures of chick embryo fibroblasts.5 Since then the antisense ODN technology has been widely used in animal sciences and as an important emerging therapeutic approach in clinical medicine.3,4 However, antisense ODN inhibition has been an under-exploited strategy for plant tissues, although the prospects for plant cells in suspension cultures to take up single-stranded ODNs was reported over a decade ago.6 In 2001, two reports from Malhó and coworker7,8 demonstrated the use of cationic-complexed antisense ODNs to suppress expression of genes encoding pollen-signaling proteins in pollen tubes from the lilly Agapanthus umbellatus. For the uptake of DNA pollen tubes represent a unique system since the growing tip is surrounded by a loose matrix of hemicellulose and pectins, exposing the plasma membrane7 and the first uptake of ODNs by pollen tubes was reported as early as 1994.9
A breakthrough in the employment of antisense ODN inhibition as a powerful approach in plant biology was recently presented through our work on intact barley leaves.10 As was illustrated by confocal microscopy and fluorescently labeled ODNs, naked ODNs were taken up through the leaf petiole and efficiently imported into the plant cell and the nucleus. The work portrayed in that study demonstrate the applicability of antisense ODN inhibition in plant biology, e.g., as a rapid antecedent to time-consuming transgenic studies, and that it operates through RNase H degradation. We employed the antisense ODN strategy to demonstrate the importance of the SUSIBA2 transcription factor11 in regulation of starch synthesis, and to depict a possible mechanism for sugar signaling in plants and how it might confer endosperm-specific gene expression during seed development.10 We also described the employment of the anitsense ODN strategy for studies on in vitro spike cultures of barley.12,13
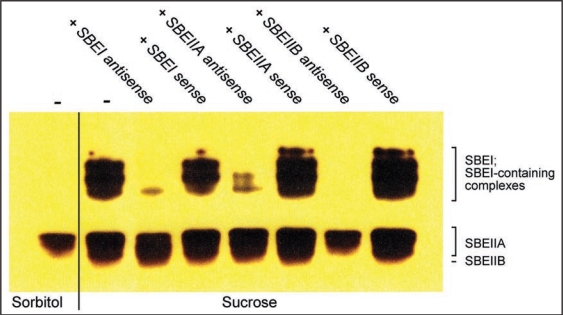
Here we present further evidence as to the value of the antisense ODN approach in plant biology by following the effects on starch branching enzyme (SBE) accumulation in barley leaves after suppression of individual SBE genes. In agreement with transcript analyses of SBE expression in barley leaves,14–16 a zymogram assay (Fig. 1) revealed that sucrose treatment of barley leaves increased the number of SBE activity bands as compared to sorbitol treatment. In the presence of antisense SBEI or SBEIIA ODNs, zymograms of sucrose-treated leaves displayed only a subset of these activities with bands in the top portion of the zymogram gel missing or diminished. With antisense SBEIIB ODN, all activity bands in the top portion of the gel as well as the lowest band were absent. Based on these data we provide a tentative annotation for the various SBE activity bands.
Figure 1.
Zymogram of starch branching enzyme (SBE) activities. Barley leaves were incubated in sorbitol or sucrose with sense or antisense SBE ODNs followed by SBE zymogram analysis.17 Antisense and corresponding sense ODNs10 were constructed as follows. SBEI (NCBI Accession number AY304541), nt 15–32; SBEIIA (NCBI Accession number AF064560), nt 1–18; SBEIIB (NCBI Accession number AF064561, nt 116–133.
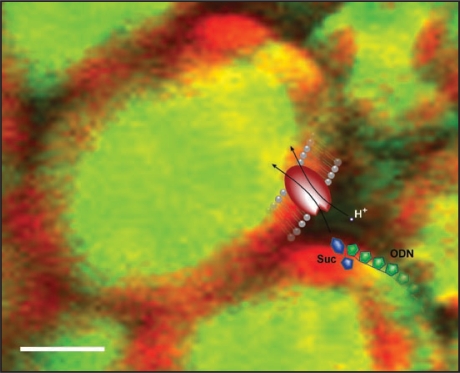
In animal experiments, naked ODNs are usually not taken up by the cells since both the ODNs and the outside of the plasma membrane carry a net negative charge. Thus the uptake of naked ODNs into barley leaf cells10 was surprising and called for an explanation. As demonstrated in our subsequent paper,13 the answer seems to be that the ODNs slip into the cells through sugar translocators as they are activated in the presence of the appropriate sugar (Fig. 2). Whether it is the structural resemblance between the sugar (deoxyribose) backbone of the ODNs and the transported sugars that allows for the ODNs to be transferred, or if other mechanisms are involved, remains to be elucidated.
Figure 2.
ODNs “piggyback” on transported sugar molecules. A pictorial representation of ODNs utilizing the sucrose translocator (SUT) to enter plant cells. The confocal microscope projection image shows uptake of fluorescently labeled ODNs (green) in barley leaves after 24 h incubation in 200 mM sucrose.10 Autofluorescence from chloroplasts is shown in red. The superimposed cartoon shows the coupled symport of H+ (white dot), sucrose (blue) and ODN (green) through the SUT (red). Scale bar, 10 µm.
Acknowledgements
This study was supported by grants from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas), and in part by U. S. Department of Energy Contract DEAC02-05CH11231 with Lawrence Berkeley National Laboratory.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/5341
References
- 1.Vanhee-Brossolet C, Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- 2.Lehner B, Williams G, Campbell RD, Sanderson CM. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 3.Gewirtz AM, Sokol DL, Ratajczak MZ. Nucleic acid therapeutics: State of the art and future prospects. Blood. 1998;92:712–736. [PubMed] [Google Scholar]
- 4.Shi F, Hoekstra D. Effective intracellular delivery of oligonucleotides in order to make sense of antisense. J Con Rel. 2004;97:189–209. doi: 10.1016/j.jconrel.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Zamecnik PC, Stephenson ML. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsutsumi N, Kanayama K, Tano S. Suppression of alpha-amylase gene expression by antisense oligodeoxynucleotide in barley cultured aleurone layers. Jpn J Genet. 1992;67:147–154. doi: 10.1266/jjg.67.147. [DOI] [PubMed] [Google Scholar]
- 7.Moutinho A, Camacho L, Haley A, Pais MS, Trewavas A, Malhó R. Antisense perturbation of protein function in living pollen tubes. Sex Plant Reprod. 2001;14:101–104. [Google Scholar]
- 8.Moutinho A, Hussey PJ, Trewavas AJ, Malhó R. cAMP acts as a second messenger in pollen tube growth and reorientation. Proc Natl Acad Sci USA. 2001;98:10481–10486. doi: 10.1073/pnas.171104598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estruch JJ, Kadwell S, Merlin E, Crossland L. Cloning and characterization of a maize pollen-specific calcium-dependent calmodulin-independent protein kinase. Proc Natl Acad Sci USA. 1994;91:8837–8841. doi: 10.1073/pnas.91.19.8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun C, Höglund AS, Olsson H, Mangelsen E, Jansson C. Antisense oligodeoxynucleotide inhibition as a potent strategy in plant biology: Identification of SUSIBA2 as a transcriptional activator in plant sugar signaling. Plant J. 2005;44:128–138. doi: 10.1111/j.1365-313X.2005.02515.x. [DOI] [PubMed] [Google Scholar]
- 11.Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansson C, Sun C, Ganeshan S, Chibbar R. Antisense ODN inhibition in in vitro spike cultures as a powerful diagnostic tool in studies on cereal grain development. Progress in Botany. 2007;68:179–190. [Google Scholar]
- 13.Sun C, Ridderstråle K, Höglund AS, Larsson LG, Jansson C. Sweet delivery—Sugar translocators as ports of entry for antisense oligodeoxynucleotides in plant cells. Plant J. 2007;52:1192–1198. doi: 10.1111/j.1365-313X.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 14.Sun C, Sathish P, Ahlandsberg S, Deiber A, Jansson C. The two genes encoding starch branching enzyme Iia and Iib are differentially expressed in barley. Plant Physiol. 1998;118:37–49. doi: 10.1104/pp.118.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mutisya J, Sathish P, Sun C, Andersson L, Ahlandsberg S, Baguma Y, Palmqvist S, Odhiambo B, Åman P, Jansson C. Starch branching enzymes in sorghum (Sorghum bicolor) and barley (Hordeum vulgare): Comparative analyses of enzyme structure and gene expression. J Plant Physiol. 2003;160:921–930. doi: 10.1078/0176-1617-00960. [DOI] [PubMed] [Google Scholar]
- 16.Mutisya J, Sun C, Rosenquist S, Baguma Y, Odhiambo B, Jansson C. Transcriptional regulation of the sbeIIb genes in sorghum (Sorghum bicolor) and barley (Hordeum vulgare): Importance of the barley sbeIIb second intron. J Plant Physiol. 2006;163:770–780. doi: 10.1016/j.jplph.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 17.Borén M, Glaring MA, Ghebremedhin H, Olsson H, Blennow A, Jansson C. Molecular and physicochemical characterization of the high-amylose barley mutant Amo1. J Cereal Sci. 2007 In press. [Google Scholar]