Abstract
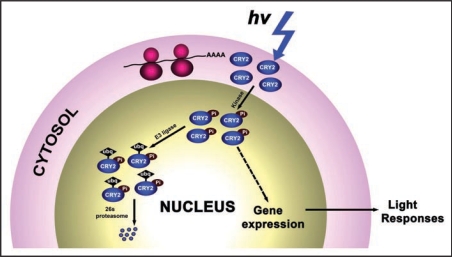
Cryptochrome 2 (CRY2) is a blue/UV-A light receptor that regulates light inhibition of cell elongation and photoperiodic promotion of floral initiation in Arabidopsis. We and others have previously shown that CRY2 is a nuclear protein that regulates gene expression to affect plant development. We also showed that CRY2 is phosphorylated in response to blue light and the phosphorylated CRY2 is most likely active and degraded in blue light. Given that protein translation (and probably chromophore attachment) takes place in the cytosol and that a photoreceptor would absorb photon instantaneously, it would be interesting to know where those inter-connected events occur in the cell. Our results showed that freshly synthesized CRY2 photoreceptor is inactive in the cytosol although it may be photon-excited, it is imported into the nucleus where the photoreceptor is phosphorylated, performs its function, becomes ubiquitinated, and eventually gets degraded (Fig. 1).1 To our knowledge, this is the first example in any organism that a photoreceptor is shown to complete its post-translational life cycle in a single subcellular compartment.
Key words: blue light, cryptochrome, ubiquitination, phosphorylation, Arabidopsis
CRY2 is Modified, Active, and Degraded in the Nucleus
Most photoreceptors are known to shuttle between different subcellular compartments in response to light. For example, rhodopsins undergo arrestin-dependent endocytosis after photoactivation;2 phototropins are released from the plasma membrane to the cytoplasm in response to blue light;3 phytochromes are imported into the nucleus from the cytosol in a light-dependent manner4–6 and cryptochromes also shuttle between the cytoplasm and nucleus.7,8 To determine whether CRY2 behaves like other photoreceptors to shuttle between different subcellular compartments, we prepared transgenic plants in the cry1cry2 mutant background that express a CRY2-GR (rat glucocorticoid receptor) fusion protein. This strategy takes advantage of well-established knowledge that a GR fusion protein is translocated into the nucleus only in the presence of a corticosteroid, such as dexamethasone. We selected transgenic lines that express the CRY2-GR fusion protein at the level not exceeding that of the endogenous CRY2 protein in the wild type plants for our study. This is because overexpression of a GR fusion protein may overwhelm the cytosolic retention machinery to cause a leak of the CRY2-GR fusion protein into the nucleus in the absence of dexamethasone. The cry1cry2 mutant shows a long hypocotyl when grown in continuous blue light and it flowers later than the wild type when grown in long-day photoperiods. Whether the CRY2-GR fusion protein can rescue the parental phenotype in the absence or presence of dexamethasone would provide a clear indication of where CRY2 performs its physiological function in the cell. We found that CRY2-GR rescued both the long-hypocotyl and delayed-flowering phenotypes only in the presence of dexamethasone, demonstrating that CRY2 functions primarily in the nucleus. Moreover, the CRY2-GR protein showed blue light-dependent phosphorylation and degradation only in the presence of dexamethasone, indicating that the modification and proteolysis of this photoreceptor also take place in the nucleus.
Can CRY2 Act in the Cytosol?
The observation that CRY2 regulate hypocotyl elongation and floral initiation in the nucleus does not necessarily exclude a possibility that CRY2 may regulate additional light responses in other subcellular compartments. A related study published recently showed that CRY1 mediates blue light inhibition of hypocotyl elongation in the nucleus. However, primary root growth and cotyledon expansion are positively regulated by the nuclear CRY1 but negatively regulated by the cytosolic CRY1.9 Interestingly, transgenic seedlings expressing the CRY2-GR fusion protein showed opened cotyledon in response to blue-light in the absence of dexamethasone. This observation may have two alternative explanations. CRY2-GR may mediate blue light stimulation of cotyledon opening in the cytosol. Alternatively, a small amount of CRY2-GR may leak into the nucleus in the absence of dexamethasone to trigger the cotyledon opening response. We have observed that some transgenic lines, which expressed the CRY2-GR fusion protein at a level higher than the lines analyzed in more details in our report, showed both short hypocotyl and early flowering phenotype in the absence of dexamethasone,1 best explained by leaks of the GR fusion protein into the nucleus in the absence of dexamethasone. In addition, CRY2 fusion proteins with an extra sequence attached to the C-terminal domain is often more active than the endogenous CRY2 (Yu, unpublished result). This may explain why a possible leak of CRY2-GR into the nucleus, at a relatively low level not apparently detectable by immunostaining or sufficient to trigger the hypocotyl inhibition response,1 might be sufficient to stimulate cotyledon opening. Whether any of the two explanations may be correct remains to be examined.
Why does CRY2 Need Continuous Blue Light for its Photoresponses?
It seems reasonable to expect that all photoreceptor molecules in the cells exposed to light can absorb photons instantaneously. Because photon-excited CRY2 is known to be phosphorylated and degraded, one may also speculate that CRY2 in blue light-treated etiolated seedlings transferred back to the dark should continue to be degraded. However, this is clearly not the case. Rather, CRY2 degradation requires continuous blue light. When etiolated seedlings were exposed to continuous blue-light, CRY2 can be degraded completely in the course of a few hours. But when light-treated plants were transferred back to the dark before completion of CRY2 degradation, most remaining CRY2 avoided the fate of destruction. This observation suggests that not all photoexcited CRY2 molecules in the nucleus are the same, and there must be at least two populations of the CRY2 pool in plants exposed to light, apparently distinguished by degradability in the dark following a light treatment. We found that phosphorylated and unphosphorylated CRY2 molecules coexist in etiolated seedlings exposed to blue light, but only the phosphorylated CRY2 was degraded in the dark. These observations argue for a scenario that CRY2 phosphorylation is the rate-limiting step for both CRY2 activity and CRY2 degradation. We hypothesized that photoexcited CRY2 is phosphorylated by a nuclear protein kinase (Fig. 1). Assuming that photoexcited CRY2 molecules are biochemically indistinguishable, the activity or abundance of the hypothetic CRY2 kinase must be light-regulated and rate-limiting. This may guarantee that only limited amount of CRY2 is phosphorylated and active at any given moment. It is known that CRY2-mediated blue-light inhibition of hypocotyl elongation is a high irradiance response that requires continuous blue light illumination.10 The CRY2-dependent flowering promotion also seems a high irradiance response (Yu, unpublished). Existence of an equilibrium of CRY2 phosphorylation may explain why these CRY2-dependent light responses requires continuous blue light exposure. This model can be directly tested upon identification of the putative CRY2 kinase.
Figure 1.
A model depicting the post-translational life cycle of CRY2. Pi, phosphate group; Ubq, ubiquitin.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/5342
References
- 1.Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin L. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell. 2007;19:3146–3156. doi: 10.1105/tpc.107.053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiselev A, Socolich M, Vinos J, Hardy RW, Zuker CS, Ranganathan R. A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron. 2000;28:139–152. doi: 10.1016/s0896-6273(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 3.Sakamoto K, Briggs WR. Cellular and subcellular localization of phototropin 1. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- 5.Nagy F, Kircher S, Schafer E. Intracellular trafficking of photoreceptors during light-induced signal transduction in plants. J Cell Sci. 2001;114:475–480. doi: 10.1242/jcs.114.3.475. [DOI] [PubMed] [Google Scholar]
- 6.Nagatani A. Light-regulated nuclear localization of phytochromes. Curr Opin Plant Biol. 2004;7:708–711. doi: 10.1016/j.pbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: Blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 8.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 9.Wu G, Spalding EP. Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc Natl Acad Sci USA. 2007;104:10013. doi: 10.1073/pnas.0705082104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]