Abstract
The macroevolution of organs and tissues in higher plants and animals may have been contingent upon the expansion of numerous gene families encoding interacting proteins. For example, there are dozens of gene families encoding actin cytoskeletal proteins that elaborate intercellular structures influencing development. Once gene family members evolve compartmentalized expression, protein isovariants are free to coevolve new interacting partners that may be incompatible with other related protein networks. Ancient classes of actin isovariants and actin-binding proteins are clear examples of such coevolving networks. Ectopic expression and suppression studies were used to dissect these interactions. In higher plants, the ectopic expression of a reproductive actin isovariant in vegetative cell types causes aberrant reorganization of the F-actin cytoskeleton and bizarre development of most organs and tissues. In contrast, overexpression of vegetative actin in vegetative cell types has little effect. The extreme ectopic actin expression phenotypes are suppressed by the coectopic expression of reproductive profilin or actin depolymerizing factor (ADF/cofilin) isovariants, but not by the overexpression of vegetative profilin or ADF. These data provide evidence for the coevolution of organ-specific protein-protein interactions. Thus, understanding the contingent relationships between the evolution of organ-specific isovariant networks and organ origination may be key to explaining multicellular development.
Key words: Arabidopsis, actin-binding proteins, actin-depolymerizing factor, profilin, actin dynamics, isovariant specificity, protein networks
A macroevolutionary context can reveal the molecular mechanisms that underlie organ and tissue development, and this is especially helpful when trying to link large gene and protein networks to particular models of multicellular development. Research attempting to connect gene family evolution, organ origination, and organ development has been limited by the complexity of both theoretical and technical approaches.1–3 Within sets of interacting gene families, we have focused on weighing the relative importance of differential gene regulation and divergence of protein isovariants as a fundamental duality often associated with the evolution of organ ontogeny.4,5 In our recent article6 we have shown how the specific interactions of coevolved members of actin and actin binding protein (ABP) isovariant families are essential for multicellular development. We used a novel approach, suppression of ectopic expression, to distinguish nonfunctional interactions from functional isovariant interactions that support normal development. In this addendum, we discuss the insights these results provide on the coevolution of networked gene families and their importance to tissue and organ development.
A significant fraction of the 30,000 to 70,000 genes in most angiosperm and vertebrate genomes encode ancient and diverse gene families.7,8,9 Since their divergence from single, common ancestral genes in simpler organisms, these duplicated family members have diversified, frequently evolving organ-and tissue-specific expression patterns. Once gene family members have evolved restricted patterns of expression, the subsets of proteins they encode may, in turn, coevolve distinct isovariant-specific interactions that are incompatible with their former networks or with concurrent but spatially separate networks in other organs. For example, there are many gene families encoding interacting proteins that comprise the actin cytoskeleton. In humans, there are four muscle and two cytoplasmic actin isovariants contributing to the cytoskeleton and cell and tissue development. In the model higher plant Arabidopsis thaliana, there are eight actin isovariants: five reproductive and three vegetative. In both animals and plants there are numerous gene families encoding differentially expressed and frequently coexpressed ABPs including actin depolymerizing factors (ADFs/cofilins) and profilins.6,10,11–14
Considering the role that the actin cytoskeleton plays in directing cellular development, it is reasonable to propose that the macroevolution of complex tissue and organ structures and the expansion and divergence of families of cytoskeletal genes and protein isovariants were contingent upon one another.1,2,3,15 We are arguing, for example, that once duplicated actin genes had evolved muscle-specific expression in early deuterostome animals or leaf-specific expression in the earliest land plants, the proteins they encoded were free to evolve novel structures distinct from their parent proteins and specific interactions with coexpressed ABPs.4,5 These coevolving sets of interacting isovariants are compartmentalized within the organs in which they are expressed. Misexpression of some of isovariants in the incorrect organ or tissue might cause dysfunctional or dominant negative phenotypes, as evidenced by research showing the functional nonequivalency of differentially expressed plant and animal actin isovariants.6,16,17
In vascular plants, the vegetative and reproductive classes of actin-based cytoskeletal systems have operated and evolved with relative independence for close to 400 million years,4,5 similar to the slightly older cytoplasmic and muscle actin systems that evolved independently in both deuterstomes and protistomes. Among the five reproductive actins in Arabidopsis, ACT1 is the most strongly expressed gene in mature pollen and germinating pollen tubes,18,19,20,21 but it is essentially undetectable in leaves or roots. In contrast, among the three vegetative actins, ACT2 is the most strongly expressed gene in leaves and roots and is not significantly expressed in pollen or pollen tubes. There are at least 16 gene families encoding ABPs in Arabidopsis22 with the profilin and ADF families being among the more thoroughly studied. The 5 profilins and 11 ADFs are similarly divided into ancient vegetative or constitutive and reproductive gene classes. For example, constitutive profilin PRF1 is expressed in all vegetative tissues, ovules, and embryos, but not in pollen, while PRF4 is expressed in mature pollen and pollen tubes.10 Likewise, ADF9 expression is restricted to vegetative tissues, while ADF7 is expressed in mature pollen and pollen tubes.11 The members of many other ABP gene families are also differentially regulated.9,22 These expression data suggest the following hypothesis: “The ancient and divergent classes of plant actin and ABP genes coevolved distinct patterns of regulation and specialized protein/protein interactions.”
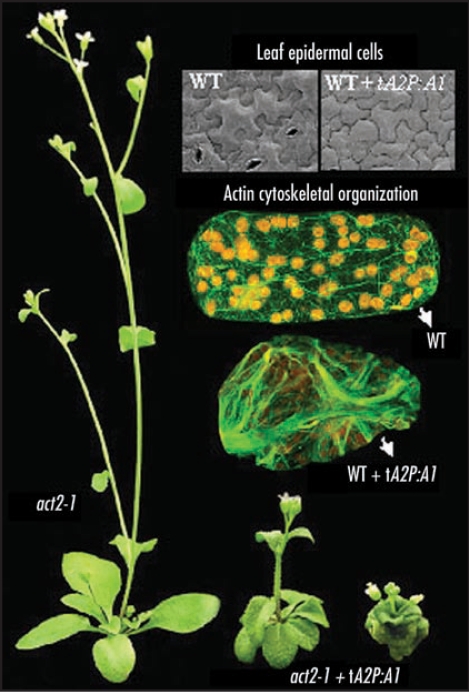
We explored ectopic expression, overexpression, and suppression to test this hypothesis for the actin cytoskeletal systems operating in Arabidopsis. We find that the transgenic misexpression of the pollen-specific reproductive actin ACT1 (tA2P::A1) in vegetative tissues of wild-type or a vegetative actin mutant (act2-1) is extremely toxic and produces severely dwarfed plants as shown in Figure 1.17 Ectopic ACT1 expression causes highly altered organization of the F-actin cytoskeleton. Every aboveground organ in these plants, including rosette and cauline leaves, inflorescence stems, sepals, petals, anthers, and pistil, are malformed. The dwarf plants contained smaller cells as shown in the scanning electron microscope image of the leaf epidermis. In contrast, the overexpression of the vegetative actin ACT2 to similar levels has little effect on plant development.17 Further evidence suggesting that the exact levels of actin expressed in vegetative tissues are not critical to development come from the fact that the act2-1 null mutant is nearly indistinguishable from WILD-TYPE. The ectopic ACT1 expression phenotypes were suppressed by the coectopic expression of the reproductive profilin or ADF.6 In particular, coectopic expression of reproductive PRF4 or ADF7 resulted in normal stature and organ development and restored normal F-actin cytoskeletal architecture to cells. On the other hand, overexpression of the vegetative ABP isovariants PRF1 or ADF9 did not suppress the ectopic ACT1 expression phenotypes.
Figure 1.
The effect of ectopic expression of reproductive ACT1 on plant morphology, cell size, and filamentous-actin organization. The act2-1 null mutant plants expressing ACT1 in vegetative tissues under control of actin ACT2 regulatory sequences (tA2P::A1) are extremely dwarf in stature (right two plants) as compared to an untransformed plant (left). The act2-1 null mutant has essentially a wild-type (WT) plant phenotype. Scanning electron micrographs reveal that the dwarf ACT1 expressing plants have smaller leaf epidermal cells. Immunolabeling of leaf cells of the dwarf plants with anti-actin antibodies reveal massive polymerization of actin into sheet-like structures (lower cell) as compared to the longitudinal arrays of fine filaments and bundles in WILD-TYPE. F-actin immunolabeling is shown in green and chloroplast autofluorescence in orange.
We hypothesized that the high-level ectopic expression of pollen actin in vegetative tissues affected actin dynamics, perhaps due to weak and in some cases inappropriate interactions among the reproductive actin and the endogenous vegetative ABPs, thus leading to excessive polymerization of F-actin and its mis-organization into star- or sheet-like structures (Fig. 1). The vegetative cytoskeletal protein network had coevolved new interactions that were incompatible with pollen actin. In support of the concept of compartmentalized coevolution, the coexpresssion of reproductive ABPs buffered the effects of ectopic ACT1 expression, allowing normal F-actin organization. Thus, the actins and ABPs had coevolved their own specific interactions in two separate developmental compartments. These data on the suppression of ectopic actin expression dissect essential class-specific interactions among the actins and ABPs and demonstrate that such interactions are required to properly organize and remodel the actin microfilament arrays necessary for normal development of different plant cell types and organs. It appears that disruption in the balance between actin and the coexpressed accessory proteins leads to deleterious consequences for cell morphology and proliferation, resulting in aberrant plant development. In the context of the fundamental duality that led us to these studies, both gene regulation and protein isovariant differences appear necessary for normal multicellular development. Both organ-specific regulation and protein structure differences must be selective factors that maintain members of the actin, profilin, and ADF/cofilins gene families in plants.
The fact that, among the hundreds of ABPs, both reproductive profilin and ADF/cofilin suppressed the ectopic ACT1 phenotype supports the view that suppression resulted from a general buffering of ACT1's toxic activities. If so, perhaps these results are typical of what will be found for many highly networked gene families with spatially distinct expression patterns. Ectopic expression or overexpression of different gene family members encoding structural proteins, metabolic enzymes, signaling proteins, and transcription factors in plants and animals frequently show strong developmental defects, implicating the importance of both regulatory and isovariant differences.16,23 –28 Mutant suppression studies within gene families are being used to test the functional roles of isovariants in multicellular development. For example, in Arabidopsis, CONSTANS (CO) encodes a transcription factor whose activity promotes the transition to flowering: the loss of CO activity results in late flowering. Many of the CONSTANS-Like COL gene family members might be expected to have redundant flower promoting activities. However, this is not the case for all COL genes. The overexpression or loss of COL9, for example, actually speeds up flowering time (unlike CO), and COL9 will not suppress CO-deficiency phenotypes.23 CO and COL9 have evolved distinct functions. In Drosophila, cytoplasmic actin isovariants cannot suppress the loss-of-flight phenotype of flies defective in the closely related flight muscle actin,16 demonstrating that small sequence differences matter in the highly networked actin cytoskeletal system. In parallel, complementation and suppression studies demonstrate that the regulatory sequences from one constitutive cytoplasmic Drosophila actin gene were essential, but the protein sequence of the particular cytoplasmic actin isovariant expressed was not important.24 Such mutant suppression studies are essential to dissecting the roles of individual gene family members and the isovariants they encode.
The next level in our understanding of the role of networked interactions in development requires in vivo evidence for the evolutionary mechanisms that have promoted gene family diversity. Testing suppression of ectopic expression among interacting families should greatly enhance our understanding of organ-specific, networked protein interactions that may be essential to the origination and maintenance of complex pathways of multicellular development. For example, our data on plant actin and ABP interactions reveal mechanisms for the origin of leaves. Leaf-like organs appear to have evolved more than once from duplicated and sterile reproductive structures (e.g., derived from sporangia) in early land plants approximately 400 mya.29–31 We have suggested that the macroevolutionary event in the angiosperm lineage that created the first leaves was contingent upon the divergence of vegetative and reproductive actin and ABP gene and protein isovariants that also occurred approximately 400 mya.4 Genome duplication and endoredundancy is relatively common in plants and nonmammalian animals, and thus, it is unlikely that gene duplication was limiting to this process. We have shown that there are specialized, coevolved interactions among actins, profilins, and ADF/cofilins that are different between the sets of proteins expressed in leaves and pollen.6 These data support the idea that the evolution of novel gene regulation and specialized isovariant interactions in the aboveground meristem may have been essential to the origin and development of the first leaves.
In summary, networked families of interacting genes and protein isovariants play key roles in multicellular development. We believe that organ origination may have been contingent upon the coevolution of these networks and the duality of differential expression and isovariant diversity within gene families. Determining the relevance of individual interactions within protein networks will require sophisticated genetic approaches. We used the suppression of ectopic isovariant expression to dissect sub-networks of actin-based systems in Arabidopsis and provided evidence for the separate coevolution of actin, profilin, and ADF/cofilin isovariants in vegetative and reproductive organs.
Acknowledgements
We would like to thank Danial Ruzicka and Gay Gragson for their critical reading of the manuscript. This research has been supported by the National Institutes of Health (GM-36397).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/5343
References
- 1.Muller GB, Newman SA. The innovation triad: An EvoDevo agenda. J Exp Zoolog B Mol Dev Evol. 2005;304:487–503. doi: 10.1002/jez.b.21081. [DOI] [PubMed] [Google Scholar]
- 2.Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128:655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Meagher RB. The impact of historical contingency on gene phylogeny: Plant actin diversity. In: Hecht M, MacIntyre R, Clegg M, editors. Evolutionary Biology. NY: Plenum Press; 1995. [Google Scholar]
- 4.Meagher RB, McKinney EC, Vitale AV. The evolution of new structures: Clues from plant cytoskeletal genes. Trends Genet. 1999;15:278–284. doi: 10.1016/s0168-9525(99)01759-x. [DOI] [PubMed] [Google Scholar]
- 5.Meagher RB, McKinney EC, Kandasamy MK. Isovariant dynamics expand and buffer the responses of complex systems: The diverse plant actin gene family. Plant Cell. 1999;11:995–1006. doi: 10.1105/tpc.11.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandasamy MK, Burgos-Rivera B, McKinney EC, Ruzicka D, Meagher RB. Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell. 2007;19:3111–3126. doi: 10.1105/tpc.107.052621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman R, Hughes AL. The temporal distribution of gene duplication events in a set of highly conserved human gene families. Mol Biol Evol. 2003;20:154–161. doi: 10.1093/molbev/msg017. [DOI] [PubMed] [Google Scholar]
- 8.Wright FA, Lemon WJ, Zhao WD, Sears R, Zhuo D, Wang JP, et al. A draft annotation and overview of the human genome. Genom Biol. 2001;2 doi: 10.1186/gb-2001-2-7-research0025. RESEARCH0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 10.Kandasamy MK, McKinney EC, Meagher RB. Plant profilin isovariants are distinctly regulated in vegetative and reproductive tissues. Cell Motil Cytoskel. 2002;52:22–32. doi: 10.1002/cm.10029. [DOI] [PubMed] [Google Scholar]
- 11.Ruzicka DR, Kandasamy MK, McKinney EC, Burgos-Rivera B, Meagher RB. The ancient subclasses of Arabidopsis ACTIN DEPOLYMERIZING FACTOR genes exhibit novel and differential expression. Plant J. 2007;52:460–472. doi: 10.1111/j.1365-313X.2007.03257.x. [DOI] [PubMed] [Google Scholar]
- 12.Meagher RB, Deal RB, Kandasamy MK, McKinney EC. Nuclear actin-related proteins as epigenetic regulators of development. Plant Physiol. 2005;139:1576–1585. doi: 10.1104/pp.105.072447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreis T, Vale R. Guidebook to the Cytoskeletal and Motor Proteins. New York: A Sambrook Tooze Publicaton at Oxford University Press; 1993. [Google Scholar]
- 14.Pollard TD. Genomics, the cytoskeleton and motility. Nature. 2001;409:842–843. doi: 10.1038/35057029. [DOI] [PubMed] [Google Scholar]
- 15.Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genom Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fyrberg EA, Fyrberg CC, Biggs JR, Saville D, Beall CJ, Ketchum A. Functional nonequivalence of Drosophila actin isoforms. Biochem Genet. 1998;36:271–287. doi: 10.1023/a:1018785127079. [DOI] [PubMed] [Google Scholar]
- 17.Kandasamy MK, McKinney EC, Meagher RB. Functional nonequivalency of actin isovariants in Arabidopsis. Mol Biol Cell. 2002;13:251–261. doi: 10.1091/mbc.01-07-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandasamy MK, McKinney EC, Meagher RB. The late pollen-specific actins in angiosperms. Plant J. 1999;18:681–691. doi: 10.1046/j.1365-313x.1999.00487.x. [DOI] [PubMed] [Google Scholar]
- 19.Huang S, An YQ, McDowell JM, McKinney EC, Meagher RB. The Arabidopsis thaliana ACT4/ACT12 actin gene subclass is strongly expressed throughout pollen development. Plant J. 1996;10:189–202. doi: 10.1046/j.1365-313x.1996.10020189.x. [DOI] [PubMed] [Google Scholar]
- 20.An YQ, Huang S, McDowell JM, McKinney EC, Meagher RB. Conserved expression of the Arabidopsis ACT1 and ACT3 actin subclass in organ primordia and mature pollen. Plant Cell. 1996;8:15–30. doi: 10.1105/tpc.8.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vitale A, Wu RJ, Cheng Z, Meagher RB. Multiple conserved 5′ elements are required for high-level pollen expression of the Arabidopsis reproductive actin ACT1. Plant Mol Biol. 2003;52:1135–1151. doi: 10.1023/b:plan.0000004309.06973.16. [DOI] [PubMed] [Google Scholar]
- 22.Meagher RB, Fechheimer M. In: The Cytoskeletal Proteome of Arabidopsis, Arabidopsis. Meyerowitz E, Somerville C, editors. Cold Spring Harbor. New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 23.Cheng XF, Wang ZY. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005;43:758–768. doi: 10.1111/j.1365-313X.2005.02491.x. [DOI] [PubMed] [Google Scholar]
- 24.Wagner CR, Mahowald AP, Miller KG. One of the two cytoplasmic actin isoforms in Drosophila is essential. Proc Natl Acad Sci USA. 2002;99:8037–8042. doi: 10.1073/pnas.082235499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim GT, Tsukaya H, Saito Y, Uchimiya H. Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:9433–9437. doi: 10.1073/pnas.96.16.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyrzykowska J, Schorderet M, Pien S, Gruissem W, Fleming AJ. Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiol. 2006;141:1338–1348. doi: 10.1104/pp.106.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, et al. Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 2006;140:1355–1366. doi: 10.1104/pp.106.077990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boucheron E, Healy JH, Bajon C, Sauvanet A, Rembur J, Noin M, et al. Ectopic expression of Arabidopsis CYCD2 and CYCD3 in tobacco has distinct effects on the structural organization of the shoot apical meristem. J Exp Bot. 2005;56:123–134. doi: 10.1093/jxb/eri001. [DOI] [PubMed] [Google Scholar]
- 29.Crane PR, Kenrick P. Diverted development of reproductive organs: A source of morphological innovation in land plants. Plant Syst Evol. 1997;106:161–174. [Google Scholar]
- 30.Floyd SK, Bowman JL. Distinct developmental mechanisms reflect the independent origins of leaves in vascular plants. Curr Biol. 2006;16:1911–1917. doi: 10.1016/j.cub.2006.07.067. [DOI] [PubMed] [Google Scholar]
- 31.Harrison CJ, Corley SB, Moylan EC, Alexander DL, Scotland RW, Langdale JA. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature. 2005;434:509–514. doi: 10.1038/nature03410. [DOI] [PubMed] [Google Scholar]