Abstract
KIR3DL1 is a polymorphic, inhibitory NK-cell receptor specific for the Bw4 epitope carried by subsets of HLA-A and -B allotypes. The Bw4 epitope is determined by the NIALR sequence motif at positions 77, 80, 81, 82 and 83 in the α1 helix. Mutation, of these positions to the residues present in the alternative and non-functional Bw6 motif, showed that the functional activity of the Bw4 epitopes of B*5101 and B*1513 is retained after substitution at positions 77, 80 and 81, but lost after substitution of position 83. Mutation of leucine to arginine at position 82, led to loss of function for B*5101 but not for B*1513. Further mutagenesis, in which B*1513 residues were replaced by their B*5101 counterparts, showed that polymorphisms in all three extracellular domains contribute to this functional difference. Prominent were positions 67 in the α1 domain, 116 in the α2 domain and 194 in the α3 domain. Lesser contributions were made by additional positions in the α2 domain. These positions are not part of the Bw4 epitope and include residues shaping the B and F pockets that determine the sequence and conformation of the peptides bound by HLA class I molecules. This analysis shows how polymorphism at sites throughout the HLA class I molecule can influence the interaction of the Bw4 epitope with KIR3DL1. This influence is likely mediated by changes in the peptides bound which alter the conformation of the Bw4 epitope.
Keywords: Human, Natural Killer Cells, MHC
Introduction
Killer cell immunoglobulin receptors (KIR), a family of inhibitory and activating HLA class I receptors, are principally expressed by NK cells. Prominent KIR are the inhibitory KIR2DL with specificity for HLA-C and the inhibitory KIR3DL with specificity for HLA-A and -B (1). KIR3DL1 is a highly polymorphic inhibitory receptor, which recognizes the Bw4 epitope carried by ~20% of HLA-A allotypes and ~33% of HLA-B allotypes. In most human populations around 50% of the HLA haplotypes encode an HLA-A and/or HLA-B allotype carrying the Bw4 epitope (2). Consequently, ~75% of people have a cognate ligand for KIR3DL1. During NK cell development the KIR gene family is expressed in variegated manner and, in combination with CD94:NKGA, an HLA-E receptor, establishes a repertoire of cells expressing different inhibitory HLA class I receptors (3). Cognate interactions between inhibitory MHC class I receptors, like KIR3DL1, and their ligands determine the extent to which mature NK cells respond to the loss of HLA class I expression that frequently accompanies cellular infection, malignancy and other trauma.
Bw4, the epitope recognized by KIR3DL1, is determined by five polymorphic positions in the helical part of the α1 domain (residues 77, 80, 81, 82 and 83) (4). In HLA-B, Bw4 bears an allotypic relationship with the Bw6 epitope carried by the majority (~67%) of HLA-B allotypes. Eight Bw4 variants are defined by polymorphism at positions 77, 80 and 81 (5). By contrast, positions 82 and 83 are invariant within the set of Bw4+ HLA-A and -B allotypes. Several studies indicate that Bw4 variants having isoleucine or threonine at position 80 are distinguished by NK cells and exhibit different clinical associations (6-8). Such effects could be mediated by different KIR3DL1 allotypes or by KIR3DS1, an activating receptor that is structurally similar to KIR3DL1 and segregates as an allele of the same locus: KIR3DL1/S1 (2).
Further complexity in the interaction of Bw4 with KIR3DL1 comes from the heterogeneous peptides bound by Bw4+ HLA-A and -B allotypes. Crystallographic structures show that KIR2DL interacts with residues 7 and 8 of the bound peptide, as well as with the segment of the α1 helix containing residue 80, for which the asparagine/lysine dimorphism determines the two KIR-recognized HLA-C specificities (9). Such overlap with the site of the Bw4 epitope and the conservation of key structural features in both the KIR2D and KIR3DL1 sequences, suggests that KIR3DL1 interacts with HLA-A and -B in the same way that KIR2DL interact with HLA-C (10). Supporting this model were observations that peptides bound to the Bw4+ allotype B*2705 do not permit interaction with KIR3DL1 if they have a charged residue at either position 7 or 8 (11). That ~25% of B*2705-binding peptides have charged residues at position 7 or 8 suggests this is no trivial effect (12, 13). And analysis of the binding of four Bw4+ A*2402 tetramers to four KIR3DL1 allotypes revealed even greater discrimination: only 6 of the 16 possible interactions occurred (14).
The goal of the study described here was to determine the contribution of variable residues in the Bw4 motif to binding of KIR3DL1. This approach also led us to show that polymorphisms outside of Bw4 epitope, and which determine peptide binding, also alter the interaction of Bw4+ HLA-B with KIR3DL1.
Materials and Methods
NK cells
PBMC were prepared from buffy coats (Stanford Blood Center) by separation on a Ficoll-Hypaque (GE Healthcare) gradient. Samples were obtained with the informed consent of the subjects. All blood collection protocols were approved by the Stanford University Institutional Review Board. Genomic DNA was prepared using the Qiagen Blood kit (Qiagen) following the manufacturer's recommendations. KIR3DL1 allele typing was performed by pyrosequencing (2). NK cell clones were derived from the PBMCs of a heterozygous donor expressing KIR3DL1*005 and *01502. PBMCs were stained with PE-Cy5-conjugated anti-CD3, PE-Cy5 conjugated anti-CD85j monoclonal antibody (mAb) specific for LILRB1, FITC- conjugated anti-CD56, PE-conjugated DX9 specific for KIR3DL1 (BD Bioscience) and propidium iodide (PI) (Sigma).
Two populations of DX9-reactive NK cells were distinguished by flow cytometry: the low binding population (DX9lo) consists of cells expressing 3DL1*005 and the high binding population (DX9hi) consists mainly of NK cells expressing 3DL1*01502, but also the small proportion of cells expressing both 3DL1*005 and 3DL1*01502. DX9lo NK cells (CD3-, CD85- CD56+ KIR3DL1*005+) and DX9hi NK cells (CD3-, CD85- CD56+ KIR3DL1*01502+) were cloned at one cell per well using a FACStar flow cytometer (Becton Dickinson). Clones were established and maintained as described (15, 16) with minor modification. Briefly, NK cell clones were cultured in Iscove's modified Dulbecco's medium (Invitrogen) containing 200 U/ml recombinant interleukin-2. At the start of culture and weekly thereafter the clones were co-cultured with 1×106/ml irradiated PBMCs from three donors. All clones had the cell surface phenotype CD3- CD85j-CD56+ KIR3DL1*005+ or KIR3DL1*01502+.
Mutagenesis of HLA-B
Inital mutagenesis of B*5101 and B*1513 was performed in a two step process as described (17). Briefly, complementary primers containing the mutation to be introduced were used in separate PCR reactions, each paired with a primer specific for the 5' or 3' end of the sequence to be amplified. The resultant amplicons were gel-purified and used as template in a second PCR reaction. This reaction used primers specific for the 5' and 3' ends of the sequence. These primers were also used to generate the full-length wild-type sequence. The final, full-length amplicons were cloned into the pEF6-V5-His expression vector (Invitrogen). Primer sequences are available on request to L. Guethlein.
Subsequent mutation of B*1513-L82R was performed using the Quik Change Multi Site Directed Mutagenesis Kit (Stratagene) following the manufacturer's recommendations. All mutations were confirmed by complete sequencing of the insert DNA.
Transfection of 221 target cells
Plasmid DNAs containing HLA-B*5101, B*1513 and mutants of these two alleles in the expression vector pEF6 were transfected into the class I deficient 221 cell line (18), by electroporation using a Gene Pulser (BioRad Laboratories) (19). Transfected 221 cells were cultured and maintained under selection by Blasticidine (5μg/ml) (Invitrogen). FITC-conjugated W6/32 mAb (eBioscience), which recognizes all HLA class I isoforms with similar avidity (20-22), was used to regularly monitor the expression of HLA-B on transfected 221 cells and to sort for high-expressing cells using a FACstar instrument. Propidium iodide was used to discriminate dead cells. Bw4-FITC and Bw6-FITC mAbs (One Lambda) were used to evaluate the effect of mutagenesis on the Bw4 and Bw6 serological epitopes.
Assay of the IFN-γ-response of NK cells to target cells
5×105 PBMCs were incubated with 221 or 221 cells transfected with HLA class I at a ratio of 1:1 in the presence of 2500U/ml IL-2 at 37° C for 14 hrs. Golgi plug (BD Biosciences) was added after one hour of incubation to inhibit IFN-γ secretion. Staining procedures were performed as described (23). In brief, following incubation with the target cell, PBMCs were stained with Dead Cell Discrimination reagent (Miltenyi), PE-Cy5 conjugated anti-CD3, PE-Cy5 conjugated anti-CD85j and PE-conjugated DX9 mAb. The cells were then fixed, permeabilized and stained with FITC conjugated anti-IFN-γ monoclonal antibody (BD Bioscience). Dead cells and cells expressing CD3 or CD85j were excluded, allowing the DX9+ NK cells to be analyzed for the intracellular accumulation of IFN-γ. The frequency of DX9+ NK cells producing IFN-γ was determined using Flowjo software. In antibody- blocking experiments, anti-class I mAb, DX17 (BD Bioscience) was used at a final concentration of 10μg/ml. Percentage IFNγ secretion was calculated using the formula: (frequency IFNγ-secreting DX9+ cells in the presence of 721.221 transfectant)/(frequency IFNγ-secreting DX9+ cells in the presence of 721.221) ×100.
Cytotoxicity assay
The cytotoxicity of NK cell clones was measured in a standard 4h 51Cr release assay using 221 cells and 221 transfectants expressing HLA class I as targets (24). In all experiments the E:T ratio was varied from 1:1 to 10:1. In antibody-blocking experiments the anti-KIR3DL1 mAb, DX9, was used at 5μg/ml final concentration. Percentage specific lysis was calculated using the formula: (Experimental 51Cr-release minus Spontaneous 51Cr-release)/(total 51Cr in target cells minus spontaneous 51Cr-release) ×100.
Results
Mutation at positions 82 and 83 of HLA-B*5101, but not mutation at positions 77 and 80, perturb the interaction with KIR3DL1
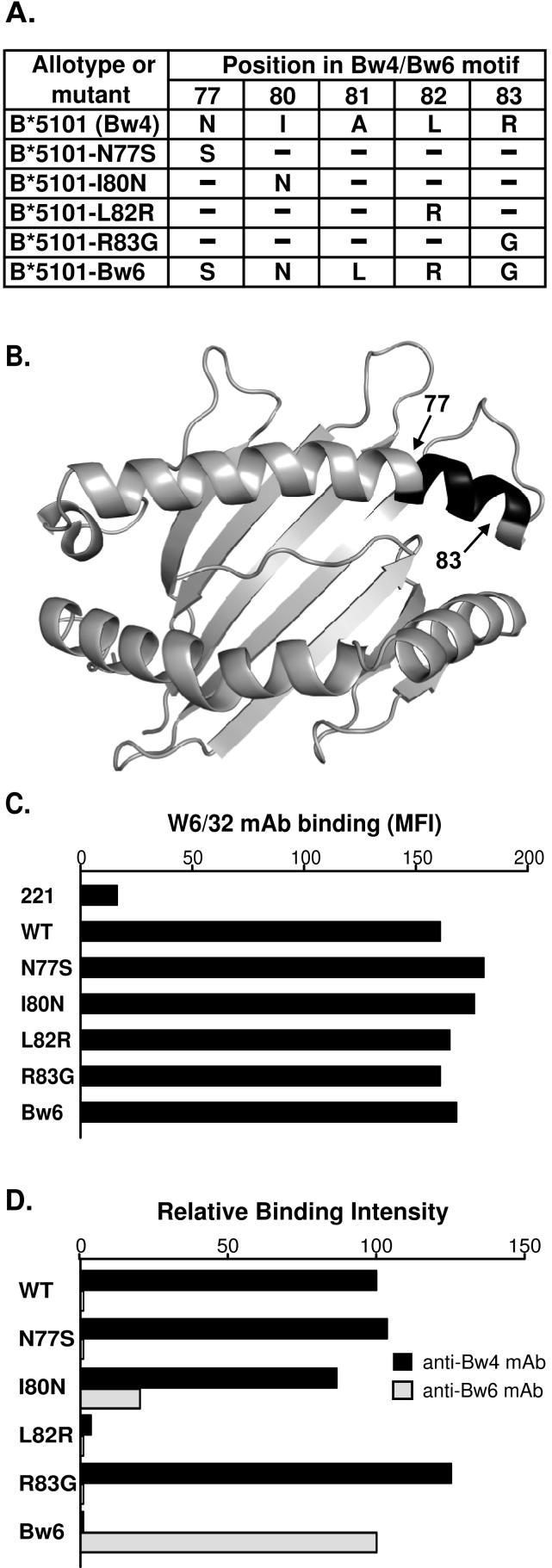
To identify which residues in the Bw4 motif of B*5101 are necessary for binding to KIR3DL1, we made point mutations at positions 77, 80, 81, 82 and 83 that distinguish Bw4 from Bw6 (Fig. 1A), the alternative motif that is not permissive for KIR3DL1 binding. A mutant B*5101 carrying the complete Bw6 motif was also made (Fig. 1B). For each mutant the corresponding residue from the Bw6 motif was introduced. Mutant cDNA were transfected into class I deficient 721.221 cells and the transfected cells assayed for their capacity to bind monoclonal antibodies specific for Bw4 (FH0007), Bw6 (FH0038) and an epitope shared by all HLA class I isoforms (W6/32). Mutants substituted at positions 77, 80, 82 and 83 were expressed similarly to B*5101 (Fig. 1C), but although the mutant at position 81 was successfully transfected, no HLA class I protein was detected at the cell surface (data not shown). Reactivity with the anti-Bw4 monoclonal antibody was retained in the mutants substituted at positions 77, 80 or 83, but lost by the position 82 mutant. Complete substitution of the Bw4 motif in B*5101 with the Bw6 motif gave a strong reaction with anti-Bw6. In contrast, four of the point mutants failed to react with anti-Bw6 and the one positive reaction, with the mutant substituted at position 80, gave a level of anti-Bw6 binding that was ~20% of that seen with the Bw6 mutant (Fig. 1D).
Figure 1. Transfected 221 cells expressing B*5101 mutants with altered Bw4 motifs.
(A) Shows the B*5101 mutants studied: four point mutants and the B*5101-Bw6 mutant, in which the Bw4 motif was replaced by the Bw6 motif. (B) Gives a top-down view of the α1 and α2 domains of B*5101 (PDB 1E28). The location of the residues defining the Bw4/Bw6 motif are shown by black shading. (C) Shows the binding of the monoclonal antibody W6/32, which is specific for a common epitope of HLA class I molecules, to class I-deficient 221 cells transfected with B*5101 or B*5101 mutants. Data are the median fluorescent intensity (MFI) as measured by flow cytometry. (Mutant B*5101-A81L was made and transfected, but was not expressed at the cell surface.) (D) Compares the binding of anti-Bw4 and anti-Bw6 monoclonal antibodies to the 221 transfected cells. The binding of Bw4 mAb or Bw6 mAb was normalized to that of W6/32 mAb binding and then calculated as relative intensity of B*5101-WT binding for Bw4 and B*5101-Bw6 binding for Bw6. Since first described (20) and characterized (21, 22) W6/32 has been extensively used and reproducibly found to be insensitive to the natural variation in HLA class I, including the Bw4/Bw6 difference. Thus the swapping of natural substitutions into the Bw4 motif of B*5101 is highly unlikely to alter the epitope recognized by W6/32, although that possibility cannot be ruled out. Experiments were performed at least 10 times with similar results. Data from a representative experiment is shown.
The capacity of the B*5101 mutants to function as ligands for four KIR3DL1 allotypes (3DL1*001, 002, 005 and 1502) was examined (Fig. 2). Populations of NK cells expressing these four allotypes were obtained from three donors as shown in Fig. 2A. Two donors expressing only one 3DL1 allotype permitted analysis of 3DL1*001- and 3DL1*002- expressing NK cells, the third donor expressed 3DL1*005 and 3DL1*1502 on different NK cell populations that were separable based on the amount of DX9 anti-KIR3DL1antibody they bound.
Figure 2. Mutations at positions 82 and 83 in the Bw4 motif of HLA-B*5101 reduce the capacity to engage KIR3DL1 and inhibit NK cells.
(A) Shows flow cytometric analysis with the anti-KIR3DL1 monoclonal antibody DX9 of NK cells from donors expressing different KIR3DL1 allotypes. NK cells from the 3DL1*005:*01502 heterozygous donor form a bimodal distribution in which the cells binding DX9 at low level express 3DL1*005 and the cells binding DX9 at high level express 3DL1*01502. The latter cells also include the small fraction of cells expressing both 3DL1*01502 and 3DL1*005; to signify this they are labeled as 3DL1*01502† cells. (B) Shows the interferon-γ response of NK cells cultured with 221 cells or 221 cells expressing wild-type (WT) or mutant HLA-B*5101. Cultures were performed in the absence or presence of the anti-HLA class I antibody DX17. Data shown here are the means from seven experiments. Statistically significant differences between mutant and wild-type are indicated by ***(p < 0.001), ** (p < 0.01), * (p < 0.05) as determined by Student's t-test. Error bars shown are SEM The antibody-blocking experiment was performed twice with similar results and data from one of those experiments is shown. (C) Shows the results of cytotoxicity assays in which NK cell clones expressing KIR3DL1*005 or KIR3DL1*01502† were incubated with 221 target cells expressing wild-type or mutant B*5101. The data were obtained at an effector to target ratio of 5:1 and are representative of five experiments.
We compared the IFN-γ response of 3DL1-expressing NK cells to class I deficient 221 cells with their response to 221 cells transfected with natural and mutant HLA class I (23). The specificity of the reactions was shown by performing the assays in the presence and absence of a blocking anti-HLA class I monoclonal antibody (DX17). Similar results were obtained with the four KIR3DL1 allotypes (Fig. 2B). The inhibitory capacity of B*5101 was unperturbed by mutation at position 77 and only slightly reduced by mutation at position 80. KIR3DL1*002 and *1502 allotypes were more affected by mutation at position 80 than the other two allotypes tested. In contrast, mutants substituted at positions 82 and 83 lost much of the inhibitory capacity for all allotypes tested. Only substitution at position 82 affected both the reactivity with KIR3DL1 and the anti-Bw4 monoclonal antibody. The effects of some combinations of KIR3DL1 allotype and HLA class I inhibitor were also examined using cytotoxicity assays, giving results similar to those observed for the cytokine response (Fig. 2C). The relative insensitivity of the inhibition to mutation at position 80 was unexpected, because dimorphism at this position determines the specificity of the HLA-C epitopes recognized by KIR2DL, and Bw4+ HLA-B allotypes with isoleucine, but not threonine, at position 80 correlate with slower progression to AIDS in KIR3DS1+ individuals infected with HIV (7, 8). To investigate this point further, we made and characterized a B*5101 mutant with alanine at position 80. This mutant also retained the inhibitory capacity of B*5101 (data not shown).
Mutation at position 83 of HLA-B*1513 perturbs interaction with KIR3DL1, but not mutation at positions 80 and 81
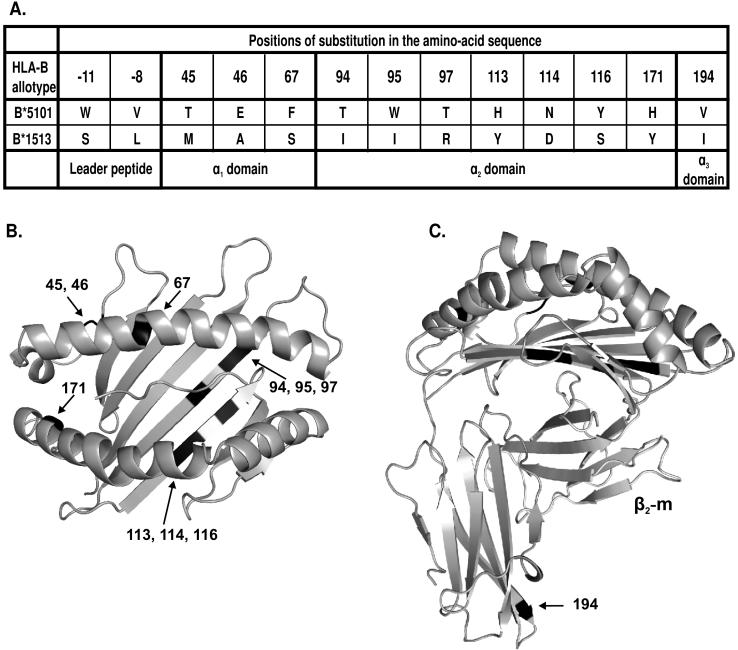
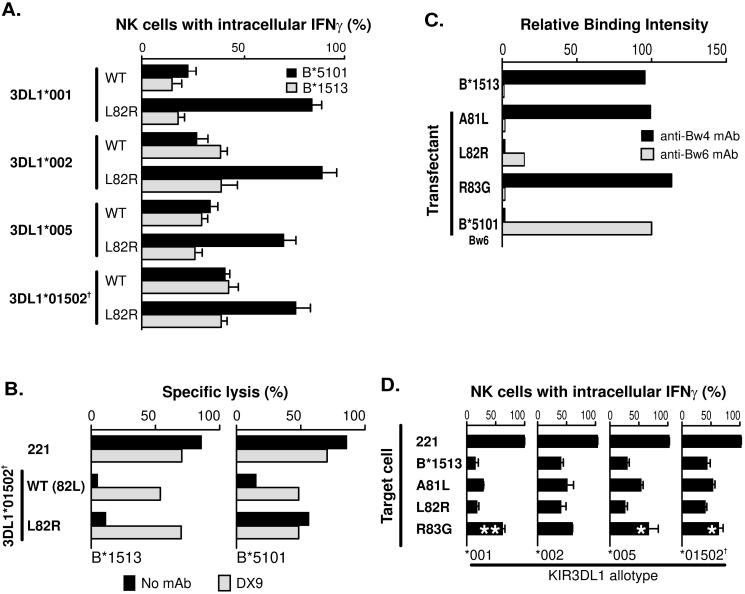
We chose the B*5101 allotype for analysis because it is common, widespread and a strong ligand for KIR3DL1 (25). Previously we reported on B*1513, a rare south east Asian allotype, which has the same Bw4 motif as B*5101 but differs elsewhere in the molecule (Fig. 3). In that study of positions 82 and 83, mutation of arginine 83 in B*1513 to glycine gave the same disruptive effect as seen here for B*5101 (26). In contrast, mutation of leucine 82 to arginine in B*1513 had no effect, whereas in B*5101 that mutation abrogated interaction with KIR3DL1. Side-by-side comparison in assays to measure the interferon-γ response (Fig. 4A) and cytotoxicity (Fig. 4B) confirmed this difference, indicating that one or more of the substitutions that distinguish B*1513 from B*5101, and which are outside of the Bw4 motif (Fig. 3), influence interaction with KIR3DL1.
Figure 3. HLA-B*5101 and B*1513 have identical Bw4 motifs but differ at thirteen positions throughout the rest of the sequence.
(A) The table shows the positions of amino-acid sequence difference between the Bw4+ B*5101 and B*1513 allotypes. (B) Gives a top-down view of the α1 and α2 domains of B*5101 (PDB 1E28). The location of the residues differing between B*5101 and B*1513 are indicated by black shading. (C) Side view of B*5101 with bound β2-m. The location of the residues differing between B*5101 and B*1513 are indicated by black shading.
Figure 4. Unlike B*5101, substitution of leucine for arginine at position 82 of B*1513 retains interaction with KIR3DL1.
(A) Compares the interferon-γ response of NK cells expressing defined KIR3DL1 allotypes cultured with 221 cells expressing wild-type (WT) or L82R mutants of HLAB*5101 or -B*1513. (B) Compares the capacity of the B*1513-L82R and B*5101-L82R mutants to engage 3DL1*01502†and inhibit the cytolytic response of NK cells to class I-deficient 221 cells. The experiment was performed three times in duplicate with similar results. Data from a representative experiment is shown in the figure. (C) Compares the binding of anit-Bw4 and anti-Bw6 mAbs to wild type and mutant B*1513. The experiment was performed eight times with similar results. Data from a representative experiment is shown. (D) Shows the capacity of wild-type and mutant B*1513 to bind different KIR3DL1 allotypes and inhibit NK cells. Data shown is the average of three experiments Statistically significant differences between mutant and wild-type are indicated by ***(p < 0.001), ** (p < 0.01), * (p < 0.05) as determined by paired t-test of means. Error bars shown are SEM. Cell surface expression level of B*1513 and its mutants was monitored with W6/32 with all transfectants expressing similar levels [data not shown].
As we had failed to obtain a B*5101 mutant at position 81, this mutation was made in B*1513. Replacing alanine 81 with leucine gave a mutant B*1513 that was well expressed at the cell surface (Fig. 4C). In functional analysis it behaved similarly to the position 80 mutant of B*5101, retaining full reactivity with monoclonal anti-Bw4 and most reactivity with KIR3DL1 (Fig. 4D). Wild-type B*1513 and its mutants at positions 81 and 82 mediated greater inhibition through 3DL1*001 than the other KIR3DL allotypes. From the combined analysis of B*5101 and B*1513, we find that arginine 83 is the one residue within the Bw4 motif that is essential for interaction with KIR3DL1. At position 83 the difference between the Bw4 and Bw6 motifs is extreme: the large positively charged arginine residue in Bw4 being replaced by the the small, neutral glycine in Bw6.
Polymorphisms at sites in the extracellular domains that are not part of the Bw4 motif influence interaction of Bw4+ HLA-B with KIR3DL1
HLA-B*5101 and B*1513 differ by 13 amino acid substitutions: two in the leader peptide, three in the α1 domain, seven in the α2 domain, and one in the α3 domain (Fig. 3). To identify which of these substitutions allowed mutation at position 82 in B*1513 (but not B*5101) to retain KIR binding, we introduced further mutations into B*1513-L82R, by replacing B*1513 residues with the substitutions present in B*5101.
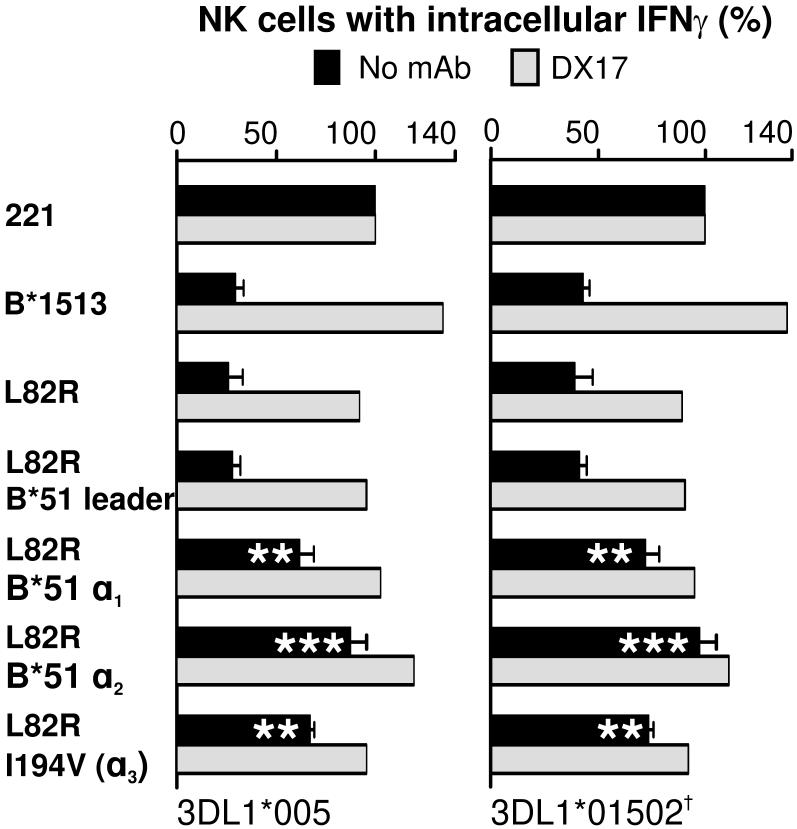
In a first set of four mutants, the leader peptide and each of the three extracelluar domains of the B*1513-L82R mutant were individually converted to the B*5101 form. Changing the leader peptide had no affect on the capacity of B*1513 to interact with KIR3DL1 and inhibit NK cell functions. In contrast, all three mutants having a B*5101 extracellular domain exhibited a reduced capacity to engage KIR3DL1 and inhibit cytokine secretion by NK cells (Fig. 5). The greatest perturbation was seen for the α2 domain conversion, with equivalent and lesser effects for the α1 and α3 domain conversions. As the α3 domain of B*1513 only differs from that of B*5101 by substitution of isoleucine for valine at position 194, these results demonstrate the contribution of isoleucine 194 to the interaction of B*1513-L82R with KIR3DL1.
Figure 5. Polymorphisms in the α1, α2 and α3 domains contribute to the differential engagement of KIR3DL1 by B*5101-L82R and B*1513-L82R.
Further mutation of B*1513-L82R generated four mutants in which the leader peptide, the α1 domain, the α2 domain, or the α3 domain, had the B*5101 sequence. The capacity of these mutants to engage KIR3DL1*005 and *01502 on NK cells and inhibit the interferon-γ response to 221 cells was measured in the presence and absence of anti-HLA class I antibody DX17. Data shown is the average of five experiments. Statistically significant differences between mutant and wild-type are indicated by ***(p < 0.001), ** (p < 0.01), * (p < 0.05) as determined by paired t-test of means. Error bars shown are SEM. The antibody-blocking experiment was performed twice with similar results and data from one of those experiments is shown.
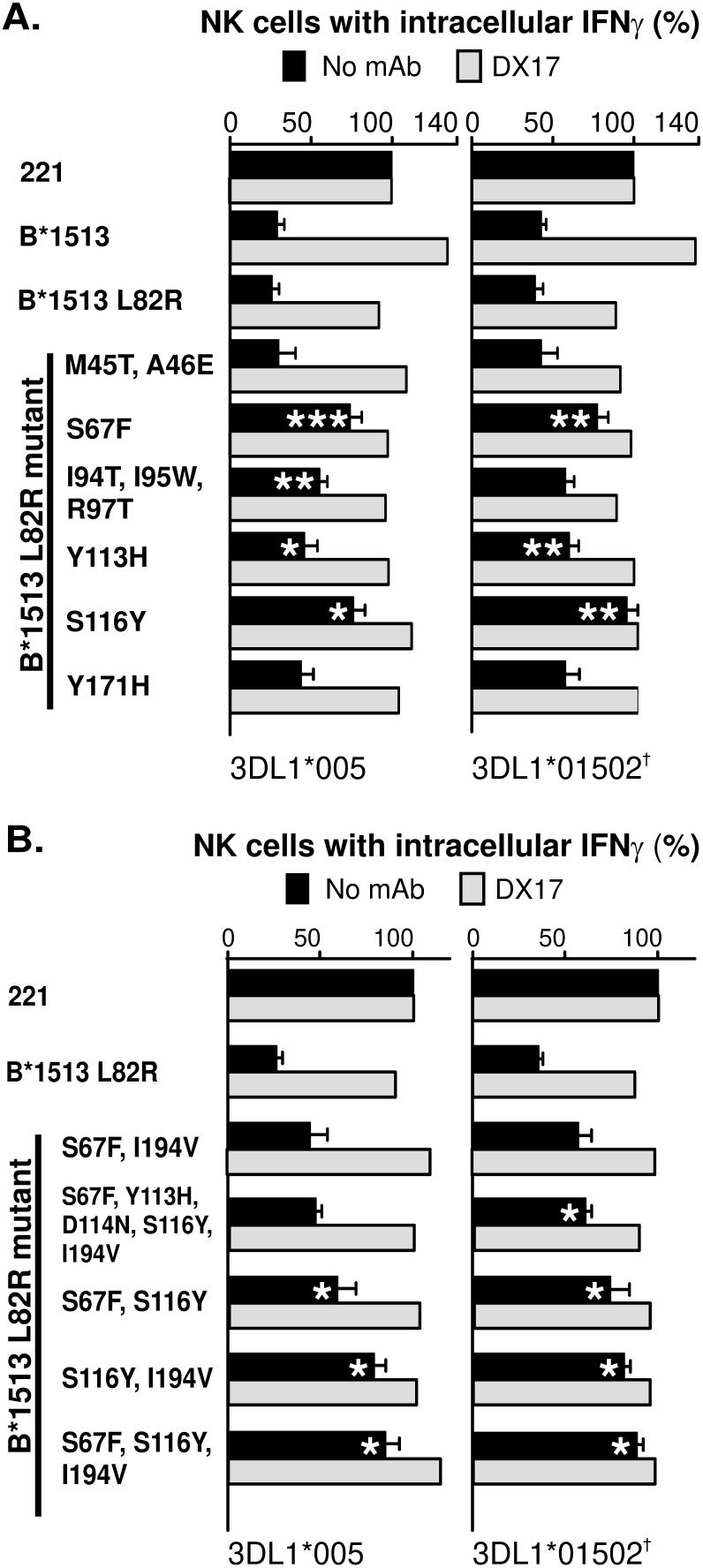
Residues 67 in α1, 116 in α2 and 194 in α3 account for most, but not all of the capacity of B*1513-L82R to engage KIR3DL1
A second set of mutants was designed to examine the effects of lone substitutions and clusters of substitutions in the α1 and α2 domains (Fig. 3). Distinguishing B*1513 and B*5101 in the α1 domain there is the pair of substitutions at positions 45 and 46 and the lone substitution at position 67. Mutation at position 67 reduced the inhibitory function of B*1513-L82R, whereas it was preserved when positions 45 and 46 were mutated (Fig. 6A). Thus, polymorphism at position 67 was responsible for the functional effect contributed by the α1 domain.
Figure 6. Differential KIR3DL1 engagement by B*5101-L82R and B*1513-L82R is principally due to residues 67, 116 and 194, but has contributions from other α2 domain residues.
Further mutation of B*1513-L82R was performed to identify individual substitutions and combinations of substitution within the α1, α2 and α3 domains that contribute to the differential engagement of KIR3DL1 by B*5101-L82R and B*1513-L82R. Assays were as described in Figure 5. (A) Shows the effects of mutation at single positions or clusters of positions within the α1 and α2 domains. (B) Shows the effect of combinations of mutations involving positions in different extracellular domains. The results are plotted as the percentage of IFN-γ secretion. Data shown here is average of three experiments. Statistically significant differences between mutant and wild-type are indicated by ***(p < 0.001), ** (p < 0.01), * (p < 0.05) as determined by paired t-test of means. Error bars shown are SEM. Cell surface expression level of B*1513 and its mutants was monitored with W6/32 with all transfectants expressing similar levels [data not shown].
Distinguishing B*1513 and B*5101 in the α2 domain is one cluster of three substitutions at positions 94, 95 and 97, another at positions 113, 114 and 116, and a lone substitution at position 171. To investigate their contribution to preserving KIR3DL1 interaction with B*1513-L82R, mutants containing these three sets of differences were constructed. The triple mutant at positions 94, 95 and 97 partially reduced the inhibition, an effect that was statistically significant for 3DL1*005 but not for 3DLl*01502 (Fig. 6A). Mutation at position 171 also had a minor effect, but did not reach significance. Because the triple mutant at positions 113, 114, and 116 failed to be expressed, we mutated these three residues individually. The mutant at position 114 failed to be expressed, whereas the mutants at positions 113 and 116 were well expressed, indicating that failed expression of the triple mutant was due to the substitution at position 114. Individual mutations at positions 113 and 116 both significantly reduced the inhibition, but the effect was greater for the 116 mutant. In conclusion, all four mutants with substitutions in the α2 domain showed diminution of inhibitory capacity (Fig. 6A). The strongest effect was due to mutation at residue 116, which reduced interaction with 3DL1 to a level comparable to that seen with the position 67 mutant. Smaller effects were observed for mutation at residues 94, 95, 97, 113 and 171. The contribution of residue 114 could not be assessed directly because of the failure of mutants at this position either to be expressed at the surface of successfully transfected cells or to be recognized by the W6/32 antibody.
To test the hypothesis that substitution at positions 67 in the α1 domain, 116 in the α2 domain and 194 in the α3 domain all contribute to the functional effect, we generated a triple mutant with all three substitutions and double mutants having the three pairwise combinations. Of these four mutants, the triple mutant showed the weakest interaction with KIR3DL1 (Fig. 6B), demonstrating that polymorphism at all three positions contributes to the differential interaction of the B*5101-L82R and B*1513-L82R mutants with KIR3DL1. Combined mutation at positions 67, 116 and 194 did not completely abrogate the interaction of B*1513-L82R with KIR3DL1, showing that one or more polymorphic residues additionally contributed to the capacity of B*1513-L82R to engage KIR3DL1 and inhibit NK cells. As an alternative approach to assess the role of position 114, for the point mutant was not expressed, we made a mutant combining substitution at positions 67, 113, 114, 116 and 194. That this mutant preserved greater interaction with KIR3DL1 than the double mutant having only the substitutions at positions 67 and 194, suggests that position 114 does not add to the effects contributed by positions 67, 116 and 194 (Fig. 6B). In summary, our results show that residues 67 in α1, 116 in α2 and 194 in α3 make a major contribution to distinguishing KIR3DL1 recognition of B*5101-L82R and B*1513. Smaller contributions are due to other residues in the α2 domain, which include 113, 171 and at least one residue from 94, 95 and 97.
Discussion
The Bw4 motif comprises three variable (77, 80 and 81) and two conserved residues (82, 83). By swap mutagenesis, in which Bw4 residues were replaced by their Bw6 counterparts, we find that none of the variable residues are essential for binding to KIR3DL1 and inhibiting NK cells. Both conserved residues are essential for the binding of Bw4+ B*5101 to KIR3DL1, but only residue 83 is essential for the binding of Bw4+ B*1513 to KIR3DL1. In a model of the binding of KIR3DL1 to HLA-B*5101 arginine 83 is shown to contact the KIR (27). Of note the isoleucine at position 80 that has been correlated with progression of HIV infection (8) is not essential for binding to KIR3DL1.
As B*5101 and B*1513 have identical Bw4 motifs, one or more of the 13 substitutions that distinguish the two allotypes was affecting the capacity of the mutant at position 82 to bind KIR3DL1. Further swap mutagenesis showed that substitutions in the leader peptide (-8 and -11) and at positions 45 and 46 in the α1 domain are not involved, and by implication neither was residue 114 in the α2 domain. Major contributions are made by positions 67 in the α1 domain, 116 in the α2 domain and 194 in the α3 domain, with smaller contributions from residues 94, 95, 97, 113 and 171. Throughout this study similar effects and trends were seen with four KIR3DL1 allotypes: 3DL1*001, *002, *005, and *1502.
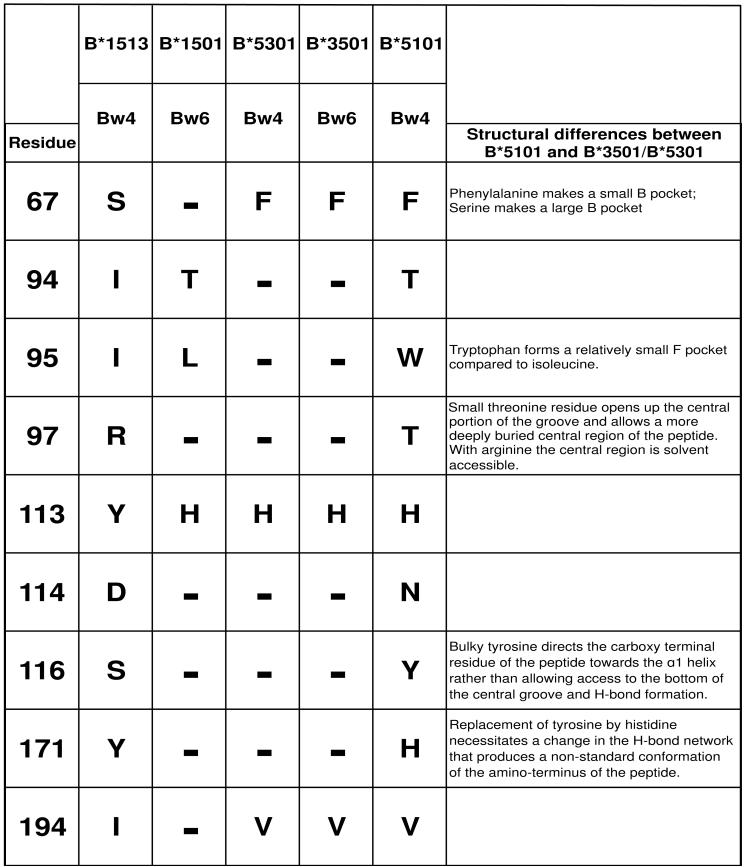
Three-dimensional structures for two complexes of antigenic peptides bound to HLA-B*5101 have been compared to structures for the serologically related HLA-B*3501 and B*5301 allotypes (27). B*5101 was seen to bind peptides in a different, non-standard manner to B*3501 and B*5301, due to an F pocket of decreased size and altered conformations for both ends of the peptide and its central region. The substitutions responsible for these differences were at positions 95, 97, 116 and 171 in the α2 domain (Fig. 7). At these four positions B*1513 has the same residues as B*3501 and B*5301, and differs from them at only two other positions (113 and 152) within the α2 domain. These similarities first predict that B*1513 binds peptide in a manner more like B*3501 and B*5301 and different from B*5101; second they point to differences in the conformation of bound peptide being responsible for the differential capacity of the L82R mutants of B*5101 and B*1513 to engage KIR3DL1. The contribution of residue 67 in α1, which affects the architecture of the B pocket also, invokes a peptide-mediated effect. In summary, this study, which shows how peptide-binding residues of the MHC class I molecule can affect the interaction of KIR3DL1, complements previous analyses showing that amino-acid sequence differences in the peptides bound by a class I molecule can create complexes that are permissive or non-permissive to KIR3DL1 binding (14, 28).
Figure 7. Changes in the B and F pockets that bind peptide anchor residues are correlated with differential KIR3DL1 engagement by B*5101-L82R and B*1513-L82R.
Shown are the nine residues that distinguish B*1513 and B*5101 and influence the binding to KIR3DL1. Also shown is their polymorphism in B*1501, B*5301 and B*3501. Identities with B*1513 are shown by a dash. B*5301 and B*3501 differ only in the Bw4/Bw6 motif. Crystallographic structures have been determined for all these allotypes, except B*1513 (27, 42-44). Although B*5101 has serological similarities with B*5301 and B*3501 it has an unusual peptide binding-site as summarised under ‗Structural differences between B*5101 and B*5301/B*3501’. At the key positions that cause these differences (67, 95, 97, 116 and 171) B*1513 is identical to B*3501. Thus the peptide binding site of B*1513 is likely to be more like those of B*5301 and B*3501, and distinct from that of B*5101.
The effect of substitution at position 194 on receptor engagement was unexpected. Residue 194 is located on the membrane-proximal end of the α3 domain at a considerable distance from the sites in the α1 and α2 domains that bind peptides and are predicted to contact KIR3DL1. Residue 194 does contribute to the site on HLA class I that binds to the LILRB1 NK cell receptor (29), raising the possibility that LILRB1 contributed to the inhibitory effects we studied. However, in the design of our experiments we deliberately excluded the participation of NK cells expressing LILRB1. What we cannot exclude, is a possible contribution from another member of the LILR family. Alternatively, the effect of substitution at position 194 may be to change the conformation of HLA-B so that it interacts less efficiently with KIR3DL1. Further experiments will be needed to distinguish these possibilities.
Structural and biochemical studies demonstrate that HLA-B*5101 is unusual compared to related HLA-B allotypes. Peptide assembly with B*5101 is slow (30), the peptides bind in a nonstandard manner (27) and with low affinity (31, 32). Our results point to the possibility that the interaction of B*5101 with KIR3DL1 is also nonstandard, as exemplified by comparison of B*5101 to B*1513. Again B*5101 seems less fit, its interaction with KIR3DL1 being highly sensitive to mutation at position 82, whereas that of B*1513 is not. This suggests that the conformation of the Bw4 epitope is significantly perturbed in B*5101, but not in B*1513, by replacement of leucine 82 with arginine. Such conformational lability in the Bw4 epitope might be a direct consequence of the low affinity binding of peptides to B*5101 and the reduced stabilization they bring to the MHC class I structure. Pertinent to this point, identical mutation of position 80 prevented expression of B*5101 but not B*1513, and the binding of different peptides to B*5101 changed the conformation of the Bw4 epitope as detected by monoclonal antibodies (33).
A further property that distinguishes B*5101 from other HLA-B allotypes is its association with Behçet's disease, a chronic and systemic inflammatory disease correlated with several genetic and environmental factors (34). NK-cells have been studied in patients with Behçet's disease (35-37) and perturbations in KIR3DL1-expressing NK cells observed (38). Polymorphisms in the genes for HLA-E and its cognate NK cell receptor CD94:NKG2A were also correlated with disease (39). Given the complementary roles that CD94:NKG2A and inhibitory KIR play in the development and function of human NK-cell repertoires (40), it is plausible that KIR may also contribute to disease susceptibility. Supporting this contention, patients with Behçet's disease are more likely to have the compound genotype of Bw4 and KIR3DL1 than matched controls (41). Thus the success, or failure, of the distinctive Bw4 epitope of B*5101 to interact with KIR3DL1 could be a factor that contributes to the incidence and progression of Behçet's disease.
Acknowledgments
This work was supported by National Institutes of Health Grants AI064520 and AI022309.
References
- 1.Bashirova AA, Martin MP, McVicar DW, Carrington M. The Killer Immunoglobulin-like Receptor Gene Cluster: Tuning the Genome for Defense. Annu Rev Genomics Hum Gene. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 2.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 3.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 4.Wan AM, Ennis P, Parham P, Holmes N. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J Immunol. 1986;137:3671–3674. [PubMed] [Google Scholar]
- 5.Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- 6.Luque I, Solana R, Galiani MD, Gonzalez R, Garcia F, Lopez de Castro JA, Pena J. Threonine 80 on HLA-B27 confers protection against lysis by a group of natural killer clones. Eur J Immunol. 1996;26:1974–1977. doi: 10.1002/eji.1830260845. [DOI] [PubMed] [Google Scholar]
- 7.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 8.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 10.Khakoo SI, Geller R, Shin S, Jenkins JA, Parham P. The D0 Domain of KIR3D Acts as a Major Histocompatibility Complex Class I Binding Enhancer. J Exp Med. 2002;196:911–921. doi: 10.1084/jem.20020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peruzzi M, Parker KC, Long EO, Malnati MS. Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J Immunol. 1996;157:3350–3356. [PubMed] [Google Scholar]
- 12.Lopez de Castro JA. HLA-B27: protraying immunodominant viral epitopes. Eur J Immunol. 2005;35:336–340. doi: 10.1002/eji.200425875. [DOI] [PubMed] [Google Scholar]
- 13.Stewart-Jones GB, di Gleria K, Kollnberger S, McMichael AJ, Jones EY, Bowness P. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur J Immunol. 2005;35:341–351. doi: 10.1002/eji.200425724. [DOI] [PubMed] [Google Scholar]
- 14.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, Carrington M, Dong T, Rowland-Jones S. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 15.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 16.Yssel H, De Vries JE, Koken M, Van Blitterswijk W, Spits H. Serum-free medium for generation and propagation of functional human cytotoxic and helper T cell clones. J Immunol Methods. 1984;72:219–227. doi: 10.1016/0022-1759(84)90450-2. [DOI] [PubMed] [Google Scholar]
- 17.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu Y, Geraghty DE, Koller BH, Orr HT, DeMars R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc Natl Acad Sci U S A. 1988;85:227–231. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. Specificity of HLA class I antigen recognition by human NK clones: evidence for clonal heterogeneity, protection by self and non-self alleles, and influence of the target cell type. J Exp Med. 1993;178:1321–1336. doi: 10.1084/jem.178.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 21.Brodsky FM, Parham P. Evolution of HLA antigenic determinants: species cross-reactions of monoclonal antibodies. Immunogenetics. 1982;15:151–166. doi: 10.1007/BF00621948. [DOI] [PubMed] [Google Scholar]
- 22.Brodsky FM, Parham P. Monomorphic anti-HLA-A,B,C monoclonal antibodies detecting molecular subunits and combinatorial determinants. J Immunol. 1982;128:129–135. [PubMed] [Google Scholar]
- 23.Draghi M, Yawata N, Gleimer M, Yawata M, Valiante NM, Parham P. Single-cell analysis of the human NK cell response to missing self and its inhibition by HLA class I. Blood. 2005;105:2028–2035. doi: 10.1182/blood-2004-08-3174. [DOI] [PubMed] [Google Scholar]
- 24.Miller RG, Dunkley M. Quantitative analysis of the 51Cr release cytotoxicity assay for cytotoxic lymphocytes. Cell Immunol. 1974;14:284–302. doi: 10.1016/0008-8749(74)90212-3. [DOI] [PubMed] [Google Scholar]
- 25.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 26.Gumperz JE, Barber LD, Valiante NM, Percival L, Phillips JH, Lanier LL, Parham P. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 27.Maenaka K, Maenaka T, Tomiyama H, Takiguchi M, Stuart DI, Jones EY. Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes. J Immunol. 2000;165:3260–3267. doi: 10.4049/jimmunol.165.6.3260. [DOI] [PubMed] [Google Scholar]
- 28.Malnati MS, Peruzzi M, Parker KC, Biddison WE, Ciccone E, Moretta A, Long EO. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 29.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 30.Sakaguchi T, Ibe M, Miwa K, Kaneko Y, Yokota S, Tanaka K, Takiguchi M. Binding of 8-mer to 11-mer peptides carrying the anchor residues to slow assembling HLA class I molecules (HLA-B*5101) Immunogenetics. 1997;45:259–265. doi: 10.1007/s002510050201. [DOI] [PubMed] [Google Scholar]
- 31.Gebreselassie D, Spiegel H, Vukmanovic S. Sampling of major histocompatibility complex class I-associated peptidome suggests relatively looser global association of HLA-B*5101 with peptides. Hum Immunol. 2006;67:894–906. doi: 10.1016/j.humimm.2006.08.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikuchi A, Sakaguchi T, Miwa K, Takamiya Y, Rammensee HG, Kaneko Y, Takiguchi M. Binding of nonamer peptides to three HLA-B51 molecules which differ by a single amino acid substitution in the A-pocket. Immunogenetics. 1996;43:268–276. doi: 10.1007/BF02440994. [DOI] [PubMed] [Google Scholar]
- 33.Takamiya Y, Sakaguchi T, Miwa K, Takiguchi M. Role of HLA-B*5101 binding nonamer peptides in formation of the HLA-Bw4 public epitope. Int Immunol. 1996;8:1027–1034. doi: 10.1093/intimm/8.7.1027. [DOI] [PubMed] [Google Scholar]
- 34.Takemoto Y, Naruse T, Namba K, Kitaichi N, Ota M, Shindo Y, Mizuki N, Gul A, Madanat W, Chams H, Davatchi F, Inoko H, Ohno S, Kimura A. Re-evaluation of heterogeneity in HLA-B*510101 associated with Behcet's disease. Tissue Antigens. 2008 doi: 10.1111/j.1399-0039.2008.01111.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Akoglu TF, Direskeneli H, Yazici H, Lawrence R. TNF, soluble IL-2R and soluble CD-8 in Behcet's disease. J Rheumatol. 1990;17:1107–1108. [PubMed] [Google Scholar]
- 36.Hamzaoui K, Ayed K, Slim A, Hamza M, Touraine J. Natural killer cell activity, interferon-gamma and antibodies to herpes viruses in patients with Behcet's disease. Clin Exp Immunol. 1990;79:28–34. doi: 10.1111/j.1365-2249.1990.tb05122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneko F, Takahashi Y, Muramatsu R, Adachi K, Miura Y, Nakane A, Minagawa T. Natural killer cell numbers and function in peripheral lymphoid cells in Behcet's disease. Br J Dermatol. 1985;113:313–318. doi: 10.1111/j.1365-2133.1985.tb02083.x. [DOI] [PubMed] [Google Scholar]
- 38.Takeno M, Shimoyama Y, Kashiwakura J, Nagafuchi H, Sakane T, Suzuki N. Abnormal killer inhibitory receptor expression on natural killer cells in patients with Behcet's disease. Rheumatol Int. 2004;24:212–216. doi: 10.1007/s00296-003-0352-x. [DOI] [PubMed] [Google Scholar]
- 39.Seo J, Park JS, Nam JH, Bang D, Sohn S, Lee ES, Park KS. Association of CD94/NKG2A, CD94/NKG2C, and its ligand HLA-E polymorphisms with Behcet's disease. Tissue Antigens. 2007;70:307–313. doi: 10.1111/j.1399-0039.2007.00907.x. [DOI] [PubMed] [Google Scholar]
- 40.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK cell repertoires towards a balance of missing-self response. Blood. 2008 doi: 10.1182/blood-2008-03-143727. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Middleton D, Meenagh A, Sleator C, Gourraud PA, Ayna T, Tozkir H, Kose AA, Azizleri G, Diler AS. No association of KIR genes with Behcet's disease. Tissue Antigens. 2007;70:435–438. doi: 10.1111/j.1399-0039.2007.00929.x. [DOI] [PubMed] [Google Scholar]
- 42.Menssen R, Orth P, Ziegler A, Saenger W. Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus. J Mol Biol. 1999;285:645–653. doi: 10.1006/jmbi.1998.2363. [DOI] [PubMed] [Google Scholar]
- 43.Smith KJ, Reid SW, Harlos K, McMichael AJ, Stuart DI, Bell JI, Jones EY. Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53. Immunity. 1996;4:215–228. doi: 10.1016/s1074-7613(00)80430-6. [DOI] [PubMed] [Google Scholar]
- 44.Smith KJ, Reid SW, Stuart DI, McMichael AJ, Jones EY, Bell JI. An altered position of the alpha 2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501. Immunity. 1996;4:203–213. doi: 10.1016/s1074-7613(00)80429-x. [DOI] [PubMed] [Google Scholar]