Abstract
In our recent paper1 we suggested a molecular explanation for the long standing observation that plants need to be mitotically active to respond to a prolonged period of low temperatures by flowering early (vernalization).2 In Arabidopsis, vernalization is associated with the epigenetic repression of the floral repressor, FLC.3–5 FLC repression is established during the low temperature treatment and is marked by the loss of chromatin marks associated with active genes and the gain of histone H3 trimethyl-lysine 27 (K27me3) at the start of transcription/translation.1 After the end of the cold treatment, this repressive modification spreads across FLC chromatin to mark the entire locus.1 In cells not undergoing mitosis, we found that FLC is repressed by low temperatures, but that this repression is only partially maintained. We concluded that DNA replication is not required for the initial response to low temperatures, but rather for the maintenance of this response. Here we discuss the implications of our observations in terms of the plasticity of chromatin modifications in plants.
Key words: trimethyl lysine 27, FLC, VIN3, bivalent domain, histone replacement
The accessibility of a gene for transcription is dependent on the structure of chromatin into which its DNA is packed. In general, the 5′ end of actively transcribed genes is associated with chromatin that is marked by high levels of H3 and H4 acetylation and H3 trimethyl-lysine 4 (K4me3), whereas chromatin marked with H3 trimethyl-lysine 27 (K27me3) is generally associated with repressed or inactive genes. Recently, promoters marked with both K4me3 and K27me3, “bivalent promoters”, were identified in pluripotent embryonic stem (ES) cells.6 These bivalently marked domains silence developmentally regulated genes, while leaving them poised for activation during differentiation. Following differentiation, bivalent promoters generally adopt either an active (K4me3) or repressed (K27me3) state, depending on cell fate.6
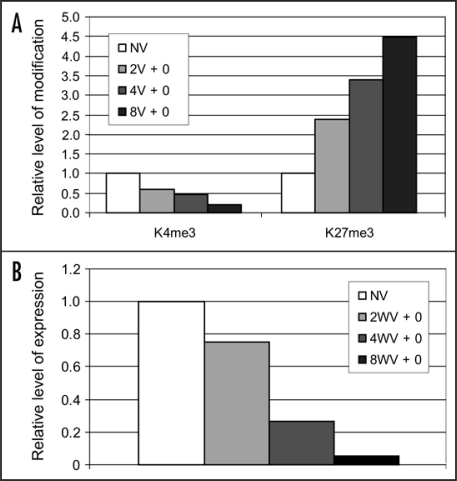
Promoters with the marks characteristic of both active and repressive chromatin also occur in plants, although it has not yet been shown whether these marks occur on the same histone or are present in different cells.7 Chromatin associated with the transcription/translation start of the FLC gene is marked with both K4me3 and K27me3.1 In contrast to the inactive, bivalently marked promoters in ES cells, FLC is expressed at high levels in non-vernalized plants. During vernalization, FLC is repressed and this is associated with a decrease in K4me3 and an increase in the level of K27me3 around the start of transcription/translation (Fig. 1A). The decrease in K4me3 and the increase in K27me3 are both proportional to the duration of the low temperature treatment, as is the level of FLC expression (Fig. 1), suggesting that the balance of these two chromatin marks controls the level of FLC transcription.
Figure 1.
The change in FLC expression and histone methylation in chromatin at the start of transcription/translation of the FLC locus is proportional to the length of the cold treatment. (A)The level of K4me3 and K27me3 in FLC chromatin was determined by chromatin immunoprecipitation assays. Quantitative polymerase chain reaction was used to quantify the immunoprecipitated FLC DNA, which was normalised to the appropriate control (for K4me3: S-ADENOSYL METHIONINE SYNTHASE; for K27me3: AGAMOUS). The data presented as a ratio of V/NV and are the mean of at least 3 independent experiments. (B) Quantitative reverse-transcription polymerase chain reaction was used to estimate the level of FLC transcripts. FDH expression was used to normalise the RNA concentration and all data points are normalised to (FLC/FDH)NV.
Vernalization-induced repression over-rides the positive effect of activators of FLC expression after the return to warmer conditions, but only in cells that are mitotically active. In these cells, the domain of K27me3 enrichment spreads from the transcription/translation start to cover the whole FLC locus.1 Mitotically-quiescent, mature leaves are unable to maintain FLC in the fully repressed state. In these leaves, the levels of K27me3 and K4me3 were restored to those in non-vernalized plants, indicating that mitotic activity is important for maintenance of repression and K27me3.
So what is the role of DNA replication in maintaining repression? Chromatin associated with active genes is enriched for the histone, H3.2 (also known as H3.3).8 This variant, which is deposited into chromatin during transcription, is enriched for covalent modifications associated with active chromatin,9–11 and at least in Arabidopsis, H3.2 does not carry K27me3.10 In contrast, histone H3.1 (also known as H3), which is deposited during DNA replication, can carry the modifications of repressive chromatin including K27me3.10,11 We suggested that histone replacement during DNA synthesis plays an important role in the conversion from H3.2 containing chromatin associated with an active FLC gene to repressive chromatin containing H3.1 marked by K27me3.1
A second gene in the vernalization response pathway, VIN3, is also associated with bivalently marked chromatin. In non-vernalized plants where VIN3 is expressed at a very low level, VIN3 chromatin is marked by K27me312 and has low levels of K4me3.13 The level of K4me3 increases during induction of VIN3 by low temperatures13 and then decreases as the gene is repressed when young growing leaves are returned to warmer conditions. In leaves that are not undergoing mitosis, VIN3 expression is not repressed on return to warmer temperatures.1 Perhaps repression of VIN3 is normally associated with the replacement of H3.2 by H3.1 during DNA synthesis allowing the balance to tip from active to repressive chromatin, just as for the repression of FLC.
The plasticity of gene expression in plants in response to changing environmental conditions is dependent on the dynamic nature of chromatin structure. Acetylation of histones is reversible thanks to the activities of a suite of histone acetyltransferase and deacetylase enzymes. Until recently, histone methylation was thought to be a stable modification, associated with long term repression of gene activity, that was removed by dilution during DNA replication or by enzymatic clipping of the histone tail followed by replacement of the histone.14,15 It has now been found that histone methylation can be removed enzymatically.16 Lysine specific demethylase 1 (LSD1) has amine oxidase activity and can remove di- and mono-methylation from H3K4.17 Homologues of LSD1 regulate flowering time in Arabidopsis.18 More recently, Jumonji (Jm°C) domain proteins have been found to remove the tri- and dimethyl group on different lysines within the H3 tail in yeast, flies and mammals.16,19 The Arabidopsis genome encodes about 20 JmjC domain containing proteins, two of which, ELF6 and REF6, play independent roles in controlling flowering time through the photoperiod and the FLC-dependent pathways respectively.20 To date none of the plant Jumonji domain proteins has been shown to have trimethyl-lysine demethylase activity.
In vernalized plants, the level of K4me3 decreased in FLC chromatin during the low temperature treatment, whether or not leaves were mitotically active. The level of K4me2 increased as K4me3 levels decreased, suggesting that loss of K4me3 occurred through active demethylation.1 Several Arabidopsis JmjC proteins show homology to JARID1, a K4me3 demethylase.21–24 Although there are no clear homologues of the K27me3 demethylases, UTX and JMJD3,25–29 our data suggest that Arabidopsis does encode K27me3 demethylase activity. In mitotically-quiescent cells K27me3 is enriched at the chromatin associated with the transcription/translation start of FLC; this mark is removed following the return to warmer conditions, concomitant with increased levels of K4me3.1 As these cells remained mitotically-quiescent even after the return to warmer conditions, this could occur by enzymatic demethylation of K27me3. In mammals, both demethylation of K27me3 and trimethylation of K4 are catalysed by components of the Mixed Lineage Leukaemia (MLL) complex;27 our data suggest that a similar complex may exist in plants. The putative K27me3 demethylase may also play a role in resetting the vernalized state in the progeny of vernalized plants.
A genome wide survey revealed that K27me3 marks almost 20% of annotated genes in ten day old Arabidopsis seedlings; many of these genes are developmentally-regulated or respond to environmental stimuli.30 Our work indicates that switching between the active and repressed states of FLC expression is a two step process involving enzymatic changes in histone methylation during the establishment phase, followed by spread of the K27me3 mark, which may be linked to histone replacement, to maintain repression. Further analysis of the chromatin modifications at other loci marked with K27me3 will demonstrate whether they too are regulated by a balance between K4 and K27 trimethylation and whether DNA replication is required to switch gene activity from the on to off state.
Acknowledgements
We thanks our colleagues Donna Bond, Diana Buzas, Chris Helliwell and Candice Sheldon for stimulating discussions and their critical reading of the manuscript.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5439
References
- 1.Finnegan EJ, Dennis ES. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol. 2007;17:1978–1983. doi: 10.1016/j.cub.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Wellensiek SJ. Dividing cells as the prerequisite for vernalization. Plant Physiol. 1964;39:832–835. doi: 10.1104/pp.39.5.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC) Proc Natl Acad Sci USA. 2000;97:418–422. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 5.Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal ARF, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 7.Saleh A, Al Abdallat A, Ndamukong I, Alvarez Venegas R, Avramova Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucl Acids Res. 2007;35:6290–6296. doi: 10.1093/nar/gkm464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Waterbourg JH. Sequence analysis of acetylation and methylation in two histone H3 variants of alfalfa. J Biol Chem. 1990;265:17157–17161. [PubMed] [Google Scholar]
- 10.Johnson L, Mollah S, Garcia BA, Muratore TL, Shabanowitz J, Hunt DF, Jacobsen SE. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of posttranslational modifications. Nucl Acids Res. 2004;32:6511–6518. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. http://epigenomics.mcdb.ucla.edu/
- 13.Finnegan EJ, Kovac KA, Jaligot E, Sheldon CC, Peacock WJ, Dennis ES. The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J. 2005;44:420–432. doi: 10.1111/j.1365-313X.2005.02541.x. [DOI] [PubMed] [Google Scholar]
- 14.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allis CD, Bowen JK, Abraham GN, Glover CV, Gorovsky MA. Proteolytic processing of histone H3 in chromatin: a physiologically regulated event in Tetrahymena micronuclei. Cell. 1980;20:55–64. doi: 10.1016/0092-8674(80)90234-2. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Jiang D, Yang W, He Y, Amasino RM. Arabidopsis relatives of the human lysine-specific demethylase 1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell. 2007;19:2975–2987. doi: 10.1105/tpc.107.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 20.Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS. Divergent roles for a pair of homologous Jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell. 2004;16:2601–2613. doi: 10.1105/tpc.104.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sewald DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, Bentley DL. Demethylatioj of trimethylated H3 lys4 in vivo by JARID1 JmjC proteins. Nature Struct Biol. 2007;14:240–242. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- 22.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument Bromage H, Tempst P, Gilliland DG, Kaerlin JWG. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Liang G, Klose RJ, Gardner KE, Zhang Y. Yeast Jhd2p is a histone H3 lys4 trimethyl demethylase. Nature Struct Biol. 2007;14:243–245. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- 24.Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protrein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 26.de Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. A histone lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 28.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquintination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 29.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18349–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of Histone H3 lysine 27 trimethylation in Arabidopsis. Plos Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]