Abstract
Flavonoid synthesis is modulated by developmental and environmental signals that control the amounts and localization of the diverse flavonoids found in plants. Flavonoids are implicated in regulating a number of physiological processes including UV protection, fertilization, auxin transport, plant architecture, gravitropism and pathogenic and symbiotic interactions with other organisms. Recently we showed that flavonoids can move long distances in plants, which may facilitate these molecules reaching positions in the plant where these processes are regulated. The localised application of selective flavonoids to tt4 mutants such as naringenin, dihydrokaempferol and dihydroquercetin showed that they were taken up at the root tip, mid-root or cotyledons and travelled long distances via cell-to-cell movement to distal tissues and converted to quercetin and kaempferol. In contrast, kaempferol and quercetin do not move long distances. They were taken up only at the root tip and did not move from this position. Here we show the movement of endogenous flavonoids by using reciprocal grafting experiments between tt4 and wild-type seedlings. These results demonstrated that to understand the distribution of flavonoids in Arabidopsis, it is necessary to know where the flavonoid biosynthetic enzymes are made and to understand the mechanisms by which certain flavonoids move from their site of synthesis.
Key words: flavonoid movement, reciprocal graft, quercetin, kaempferol, Arabidopsis thaliana, fluorescence, aglycone
Flavonoids are plant secondary metabolites made by the phenylpropanoid pathway. The central biosynthetic pathway is known and in Arabidopsis most of the enzymes in flavonoid synthesis are encoded by single copy genes.1 The isolation of mutants with defects in the genes encoding these flavonoid biosynthetic enzymes has allowed researchers to understand the biochemical complexity of flavonoid synthesis and their biological roles. Flavonoid synthesis is more complex in other species, such as legumes, which produce a greater diversity of flavonoid molecules, and in which gene families encode the key enzymatic branch points of the pathway.2,3
The functions of flavonoids were demonstrated using genetic approaches that blocked flavonoid synthesis in Arabidopsis and other species. In Arabidopsis, flavonoids play important roles in UV protection4 and regulate auxin transport and dependent physiological processes, such as gravity responses,5–7 and lateral root formation.8 In petunia, maize and tomato, pollen without flavonoids is infertile and this phenotype is reversed by flavonoid addition.9–11 However, the enigma of why flavonoid-deficient Arabidopsis seedlings are fertile has not been resolved.12 Flavonoids appear to interact with Multidrug resistance (MDR)/P-glycoproteins (PGP)/ABC-Type B proteins7 that transport auxin, regulate phosphatases and kinases, and may have regulatory roles as scavengers of reactive oxygen species (reviewed in ref. 13). These results are consistent with a diversity of important functions for flavonoids in plants that require careful control of flavonoid synthesis and localization.
We have explored the possibility that flavonoid accumulation in specific locations is also modulated by movement of early intermediates of the flavonoid pathway. Long-distance movement of secondary metabolites is largely unexplored but potentially has profound developmental effects. Grafting experiments conducted in the early 1900s suggested that alkaloids move from the site of manufacture (the root) to the aerial tissue.14 More recent grafting experiments showed that root synthesised metabolites, perhaps carotenoids, regulate shoot development,15,16 flowering inducers travel long distances,17 and phytohormones are translocated (reviewed in ref. 18).
We recently showed that flavonoids moved long distances in Arabidopsis using several approaches.19 The roots of Arabidopsis seedlings grown in complete darkness do not accumulate flavonoids5 since expression of early genes encoding enzymes of flavonoid biosynthesis are light dependent.20 Yet, flavonoids accumulate in root tips of seedlings with light-grown shoots and light-shielded roots, consistent with shoot-to-root flavonoid movement. Using fluorescence microscopy, a selective flavonoid stain (diphenyl boric acid 2-amino ethyl ester [DPBA]), and localised aglycone application to transparent testa mutants, we showed that flavonoids accumulated in tissues distal to the application site, indicating that early intermediates in the flavonoid pathway can move long distances. This was confirmed by time-course fluorescence experiments and HPLC. Flavonoid applications to root tips resulted in basipetal movement in epidermal layers, with subsequent fluorescence detected 1 cm from application sites after 1 h. Flavonoid application mid-root or to cotyledons showed movement of flavonoids toward the root tip mainly in vascular tissue. Naringenin, dihydrokaempferol and dihydroquercetin were taken up at the root tip, mid-root or through cotyledons and travelled long distances via cell-to-cell movement to distal tissues followed by conversion to quercetin and kaempferol. In contrast, kaempferol and quercetin were only taken up at the root tip. Uptake of flavonoids at the root tip was inhibited by glybenclamide, a specific inhibitor of ABC type transporters21 suggesting a possible role for transporters of this class in the movement of flavonoids.
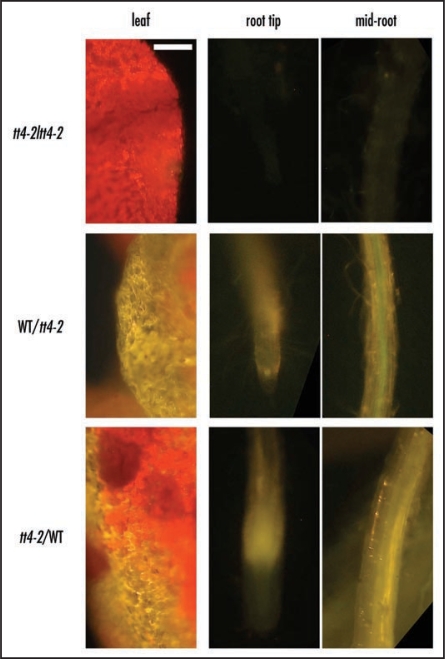
To show that endogenous flavonoids are capable of long distance movement, we performed reciprocal butt grafting between tt4 and wild-type seedlings.22 In these experiments we asked whether flavonoids moved from wild-type tissues to flavonoid-deficient tissues of tt4. DPBA fluorescence detection was used to detect flavonoid movement into tt4 tissues.19 Seedlings were grafted and grown on filter paper in Petri dishes for 8 d. The seedlings were then transferred to equal parts sand, perlite and vermiculite to avoid the possibility of uptake of pre-existing flavonoids that may be natural soil components. After 14 d, the seedlings were stained with DPBA. When tt4 roots were grafted to tt4 shoots, the samples showed dim greenish autofluorescence in roots and only red chlorophyll fluorescence in the shoot. When either tt4 roots or shoots were grafted onto wild-type shoots or roots, respectively, the tt4 tissues showed bright yellow DPBA fluorescence (Fig. 1). The results of these experiments clearly showed that endogenous flavonoids moved across the graft to the reciprocal tissue. The flavonoid movement is specific to certain tissues, as flavonoids are not transported into the seeds developing on tt4 shoots grafted on wild-type roots, which retain the transparent testa phenotype. Although flavonoids clearly travelled from wild-type root tissue to mutant shoots, they were not capable of complementing the seed colour defect of tt4. In addition, adding naringenin to tt4 plants either to the media, or to the soil, also did not complement the seed colour phenotype in tt4 (data not shown). Recent research by Hsieh and Huang23 may account for this inability to complement seed colouration, as flavonoids in the Brassicaceae end up in the pollen coat rather than the testa. The testa tissue derives from ovular tissue,24 and thus is maternal in origin.
Figure 1.
Grafting shows flavonoid movement occurs across grafts. Reciprocal grafting between wild type and tt4 indicated flavonoid movement from the flavonoid producing tissue to the chalcone synthase-deficient tissue. The order of the graft is indicated by the left legend as aerial tissue over root tissue in the graft. Micrographs are DPBA stained tissue excited with 488 nm wavelength. The tt4/tt4 control graft shows no flavonoids are present, even though a wound has occurred on the leaf which generally exacerbates flavonoid fluorescence. Scale bar = 100 µm. Green fluorescence is from kaempferol and gold from quercetin. The red fluorescence is from chlorophyll.
The complexity of the flavonoid biosynthetic pathway and the large number of modified flavonoids that can be made through the complex series of glycosylation reactions suggests that distinct flavonoid molecules may have unique function. To fully understand these molecules, it is necessary to dissect the synthesis pathways for these glycosylated flavonoids. Two unnamed flavonoid glycoside mutants isolated in 1998,25 have profound developmental phenotypes, supporting this hypothesis. These mutations resulted in whorled cauline leaves on inflorescences and double the number of rosette leaves. Our lab is in the process of determining if other phenotypes exist in flavonoid mutants.
A critical feature of the observations of flavonoid movement is understanding the biological context of this movement. First, a number of recent studies reported physiological functions of flavonoids in roots, ranging from modulation of auxin transport and root gravitropism,5–8 to nodulation3 and root branching,8 while it is clear that flavonoid synthesis is absent in dark-grown seedlings.5 Yet, for flavonoids to function in roots of plants grown in soil, the light signal and/or flavonoid precursors must travel to the roots. Additionally, transient flavonoid accumulation has been reported in roots reoriented relative to the gravity vector.6 For flavonoids to transiently accumulate at the root tip (at 2 hours after reorientation) and to return to lower levels (within 2 additional hours), suggests that more than flavonoid synthesis is regulated. Perhaps this transient flavonoid accumulation requires localized enzyme activation and transport mechanisms. As flavonoid transport is inhibited by a compound that blocks ABC transporters, which include the newly identified auxin transporters of the MDR/PGP class, perhaps there are connections between flavonoid and auxin transport that allow this transient accumulation. A more detailed understanding of this role of flavonoid movement in controlling plant development awaits additional experimentation.
Acknowledgements
The Australian Research Council Centre of Excellence for Integrative Legume Research Project ID CEO348212 and the Biotechnology Resource Centre, the Australian National University (to M.A.D.) and the United States Department of Agriculture 2006-03406 (to G.K.M.).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5440
References
- 1.Winkel Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki T, Akashi T, Ayabe S. Flavonoids in leguminous plants: structure, biological activity, and biosynthesis. J Plant Res. 2000;113:475–488. [Google Scholar]
- 3.Wasson AP, Pellerone FI, Mathesius U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by Rhizobia. Plant Cell. 2006;18:1617–1629. doi: 10.1105/tpc.105.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Ou Lee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buer CS, Muday GK. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell. 2004;16:1191–1205. doi: 10.1105/tpc.020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buer CS, Sukumar P, Muday GK. Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol. 2006;140:1384–1396. doi: 10.1104/pp.105.075671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis DR, Miller ND, Splitt BL, Wu G, Spalding EP. Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis Multidrug Resistance-Like ABC transporter genes. Plant Cell. 2007;19:1838–1850. doi: 10.1105/tpc.107.051599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 2001;126:524–535. doi: 10.1104/pp.126.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coe EH, McCormick SM, Modena SA. White pollen in maize. J Hered. 1981;72:318–320. [Google Scholar]
- 10.Mo Y, Nagel C, Taylor LP. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA. 1992;89:7213–7217. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ylstra B, Busscher J, Franken J, Hollman PCH, Mol JNM, van Tunen AJ. Flavonols and fertilization in Petunia hybrida—localization and mode of action during pollen tube growth. Plant J. 1994;6:201–212. [Google Scholar]
- 12.Ylstra B, Muskens M, van Tunen AJ. Flavonols are not essential for fertilization in Arabidopsis. Plant Mol Biol. 1996;32:1155–1158. doi: 10.1007/BF00041399. [DOI] [PubMed] [Google Scholar]
- 13.Peer WA, Murphy AS. Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 2007;12:556–563. doi: 10.1016/j.tplants.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Waller GR, Nowacki EK. Alkaloid biology and metabolism in plants. New York: Plenum Press; 1978. pp. 121–141. [Google Scholar]
- 15.Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, Leyser O. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003;17:1469–1474. doi: 10.1101/gad.256603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Norman JM, Frederick RL, Sieburth LE. BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr Biol. 2004;14:1739–1746. doi: 10.1016/j.cub.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 17.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signalling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 18.Weyers JDB, Paterson NW. Plant hormones and the control of physiological processes. New Phytol. 2001;152:375–407. doi: 10.1046/j.0028-646X.2001.00281.x. [DOI] [PubMed] [Google Scholar]
- 19.Buer CS, Muday GK, Djordjevic MA. Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol. 2007;145:478–490. doi: 10.1104/pp.107.101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelletier MK, Shirley BW. Analysis of flavanone3-hydroxylase in arabidopsis seedlings. Plant Physiol. 1996;111:339–345. doi: 10.1104/pp.111.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forestier C, Frangne N, Eggmann T, Klein M. Differential sensitivity of plant and yeast MRP (ABCC)-mediated organic anion transport processes towards sulfonylureas. FEBS Lett. 2003;554:23–29. doi: 10.1016/s0014-5793(03)01064-0. [DOI] [PubMed] [Google Scholar]
- 22.Turnbull CGN, Booker JP. Leyser HMO (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 32:255–262. doi: 10.1046/j.1365-313x.2002.01419.x. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh K, Huang AHC. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell. 2007;19:582–596. doi: 10.1105/tpc.106.049049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debeaujon I, Léon-Kloosteziel KM, Koornneef M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 2000;122:403–413. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham TL. Flavonoid and flavonol glycoside metabolism in Arabidopsis. Plant Physiol Biochem. 1998;36:135–144. [Google Scholar]