Abstract
This is an addendum to our recent paper published in The Plant Journal (52:352–61). The major findings were: (1) trichomes on the leaves of gl3-sst sim double mutants developed as large multi-cellular clusters whereas wild type trichomes are composed of single cells; (2) ectopic CYCD3;1 expression in gl3-sst trichomes also resulted in trichome cluster formation; and (3) that GL1 expression is prolonged in the gl3-sst sim trichome clusters. This addendum shows that ectopic CYCD3;1 expression in gl3-sst also enhanced GL1 expression. An analysis of the GL1 promoter found two overlapping potential E2F binding sites in a region of the promoter known to be essential for GL1 function. This finding indicates that GL1 may be directly regulated by the activity of a CYCD3/CDKA complex that phosphorylates E2F-RB bound to the GL1 promoter.
Key words: plant cell cycle, endoreduplication, glabra1, plant development
Introduction
The development of Arabidopsis trichomes is being used as a model to address basic biological questions concerning control of cell fate and differentiation.1–4 On the leaves, trichomes are composed of large unicellular structures containing a stalk and three to four branches. These are the first cells to terminally differentiate on young leaf primordia, and, like many differentiated plant cells, they undergo endoreduplication (excess DNA replication without cell division). Over the past twenty years a core of transcription factors have been identified that are required for the trichome cell fate. These include GLABROUS1 (GL1; a R2R3 MYB), TRANSPARANT TESTA GLABRA1 (TTG1; a WD40 repeat containing protein) and the semi-redundant pair of genes GLABRA3 and ENHANCER OF GLABRA3 (GL3 and EGL3; bHLH proteins).5–8 Mutations in any of the three classes of genes result in a loss of trichome initiation on most plant epidermal surfaces. Several studies indicate that these factors interact, and likely form a transcriptional complex that regulates genes required for trichome formation.6,8,9
Whereas double gl3 egl3 mutants lack trichomes, plants deficient in GL3 have more slender less branched trichomes that exhibit reduced endoreduplication (8–16C vs. 32–64C).8,10 Interestingly, a special mutant allele of GL3 called gl3-shapeshifter (gl3-sst) has been identified that induces enlarged variably shaped and branched trichomes that exhibit extremely enhanced endoreduplication (over 200C).9,11 These mutants also develop fewer trichomes. The mutant gl3-sst allele contains a single base pair change that converts a leucine to a phenylalanine residue in the region of the GL3 peptide that interacts with GL1. Yeast two hybrid analyses have shown that the substitution results in decreased interaction between GL1 and gl3-sst polypeptides9. A model explaining the gl3-sst phenotype posits that the reduced interaction between GL1 and gl3-sst results in fewer cells acquiring a threshold level of activator complex needed to activate genes required for commitment to the trichome fate. The extra endoreduplication and cell expansion of those few trichomes that do develop are thought to be due to the inability of the altered GL1/gl3-sst containing complex to activate genes needed to limit trichome growth during early phases of trichome differentiation. Indeed, it has been found that the expression of TRIPTYCHON (TRY), which functions to limit trichome expansion and endoreduplication, is decreased in gl3-sst trichomes.12,13
A genetic screen was conducted to identify additional genes that might play a role in the gl3-sst phenotype. This screen resulted in the identification of a mutant that exhibited large clusters of trichome cells with apparent meristematic activity.11 The clusters were shown to be derived from individual trichome precursors that repeatedly underwent anticlinal and periclinal cell divisions. Outcrossing of the mutant to a wild type plant resulted in an F2 population containing a new mutant phenotype. This mutant exhibited trichomes that appeared somewhat normal, but on closer inspection were found to be composed of multiple cells. This phenotype was identical to that described for the SIAMESE (SIM) mutant.14 Allele testing confirmed that the mutants were allelic. SIM has been cloned and found to encode a protein related to proteins that inhibit cyclin D activity.15 Further, it was shown that SIM protein interacts with both CYCDs and Cyclin Dependent Kinase A (CDKA).
CYCDs are thought to be important for both G1 to S phase and G2 to M phase transitions.16 In wild type trichomes, it has been shown that ectopic expression of CYCD3;1 induces the formation of sim-like multicellular trichomes that have fairly normal morphology.17 To test if the clusters found in gl3-sst sim could be due to extra CYCD activity, CYCD3;1 was ectopically expressed via the GLABRA2 promoter (GL2; drives high level of expression in trichomes18) in the gl3-sst mutants.11 The resulting plants formed trichomes that roughly phenocopied those seen on gl3-sst sim double mutants. However, there were differences. For example, the trichomes on gl3-sst CYCD3;1 plants exhibited a delay in cell division and the resulting clusters contained more branched trichomes than seen in gl3-sst sim clusters.
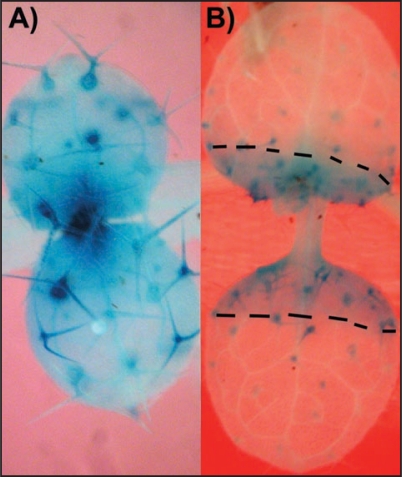
GL1 also appears to play a role in the formation of the clusters. GL1 is normally expressed throughout young leaf primordia and then is up regulated in developing trichomes (Fig. 1A).19 A zone of GL1 expression in young leaves persists but abruptly disappears once a leaf reaches a certain size (Fig. 1B). In addition, GL1 expression normally shuts down as trichomes develop. In gl3-sst sim, high GL1 expression was found to persist in mature trichome clusters.11 This high level of GL1 expression required the loss of SIM activity as gl3-sst trichomes showed more normal levels of GL1 expression. For this addendum we wished to explore the status of GL1 expression in gl3-sst CYCD3;1 trichomes. The key question was whether or not altering the cell cycle through ectopic CYC3;1 expression can alter GL1 expression.
Figure 1.
GL1::GUS expression in young Arabidopsis shoots. (A) GL1::GUS expression in a young Arabidopsis seedling. Intense staining is seen in the trichomes and apex region, whereas lighter more diffuse staining is seen throughout the leaf surface. (B) Slightly older seedling showing abrupt loss of diffuse surface staining and reduced expression in trichomes. The GL1::GUS construct is described in ref. 19 and is available from the ABRC stock center (CDC-364 plasmid or CS8850 seeds).
In Marks et al., (2007) real time qPCR was used to compare levels of GL1 expression between young shoot apexes, where GL1 expression is highest in wild type (see Fig. 1A), to young developing leaves where GL1 expression rapidly declines (Fig. 1B). This analysis showed that GL1 expression decreased over a 1000-fold from apex to leaf. Similar results were obtained for the individual gl3-sst and sim mutants. However, as stated above similar levels of GL1 expression were found in the apexes and young leaves of gl3-sst sim plants. In the gl3-sst sim leaves, most of the GL1 mRNA was likely derived from the trichome clusters because almost all of the GUS staining in GL1:: GUS containing gl3-sst sim plants was seen in the trichome clusters. The real time qPCR experiment has been repeated to compare GL1 mRNA levels in the apexes and young leaves of wild type, gl3-sst sim, and gl3-sst GL2::CYCD3;1 plants.
New Findings
As shown in Table 1, GL1 expression in the apex tissue was similar in all three genotypes. And as previously shown, GL1 expression falls dramatically in wild type young leaf tissue (declined by 490 fold), but holds fairly steady in gl3-sst sim (declined by only 1.5 fold). The expression in gl3-sst GL2::CYCD3;1 young leaves was reduced by 19.6 fold. This level of reduction is much less than seen for wild type or in the previous analysis of gl3-sst alone. Thus, the increase in expression is likely due to elevated CYCD3;1 expression. This result suggests that there is a linkage between progression through the cell cycle and GL1 expression in developing trichomes.
Table 1.
Relative levels of GL1 and HSP70 expression4
| Gene | Tissue | Relative Quantity1 | Shoot over Leaf |
| GL1 | Col shoot2 | 1.000 ± 0.063 | 490 |
| GL1 | Col leaf3 | 2.04 × 10−3 ± 0.045 × 10−3 | |
| GL1 | sstsim shoot | 1.586 ± 0.093 | 1.5 |
| GL1 | sstsim leaf | 1.043 ± 0.109 | |
| GL1 | sst cycD3;1 shoot | 2.666 ± 0.701 | 19.6 |
| GL1 | sst cycD3;1 leaf | 0.136 ± 0.011 | |
| HSP705 | Col shoot2 | 1.000 ± 0.018 | 1.09 |
| HSP70 | Col leaf | 0.915 ± 0.026 | |
| HSP70 | sstsim shoot | 1.195 ± 0.099 | 1.2 |
| HSP70 | sstsim leaf | 1.028 ± 0.051 | |
| HSP70 | sst cycD3;1 shoot | 1.578 ± 0.096 | 1.85 |
| HSP70 | sst cycD3;1 leaf | 0.852 ± 0.082 |
All quantities were normalized to the level of expression obtained for Col shoot. GL1 and HSP70 values were normalized separately. Mean ± standard deviation values are shown derived from 2 replicates of each tissue.
All young shoot tissue contained just the apex and emerging leaves that lacked petioles.
Third leaves were dissected from seedlings just at the stage when the leaves were beginning to develop petioles.
All reaction conditions and primers are described in Marks et al., 2007.
The HSP70 gene was used an internal control of mRNA integrity, as previous analyses have shown that the expression of this gene varies little from tissue to tissue under normal growth conditions.
E2F and Gene Expression in Trichomes
One of the key targets of CYCD/CDKA activity is the RETNOBLASTOMA RELATED (RBR) protein.20,21 RBR binds to the transcription factor E2F to repress its activity. Phosphorylation of RBR by CDKA reduces the interaction between RBR and E2F.16 After the release of of E2F from phosphorylated RBR, E2F is free to activate the expression of target genes, many of which have roles in DNA replication.22 The interaction between E2F and RBR also can result in direct inactivation of the expression of gene targets.23 This is thought to occur when E2F recruits RBR to target genes where interactions between RBR and histone deacetylases can induce localized heterochromatin formation to silence gene expression. There is some evidence that E2F can play a role in trichome gene expression. Ramirez-Parra and Gutierrez (2007) have shown via GUS reporter constructs that the FASCIATA1 (FAS1) gene, which encodes a component of the Chromatin Assembly Factor 1, is expressed in trichomes. Further, they were able to show that mutations in one of two E2F binding sites in the FAS1 promoter enhanced gene expression, whereas mutations the nearby second E2F binding site repressed gene expression.24
GL1 Expression and the Cell Cycle
In developing trichomes, GL1 expression is greatest during the same interval of time that endoreduplication occurs.10,19 This is a period during which CYCD/CDKA activity would be involved in promoting this activity. Thus, if GL1 expression is directly modulated by E2F, then the rise and fall in CDKA activity during endoreduplication could play a role in GL1 expression. This could occur either through direct up regulation via E2F activation or via repression through chromatin remodeling.
In a previous analysis, promoter sequences responsible for GL1 expression and function were identified.19 Interestingly, a 153 bp interval starting 1028 bp downstream of the stop codon was essential for GL1 gene function. Genomic fragments lacking this region failed to restore trichome development in gl1 mutants. An analysis of the sequences 1000 bp upstream of the start codon and over 2800 bp downstream of the stop codon revealed three potential E2F binding sites. One of these is in the large second intron of the gene. This site is likely not responsible for regulating GL1 expression as this site is not present in the GL1::GUS construct that revealed reduced activity in mature wild type trichomes and high GL1 expression in gl3-sst sim clusters. However, at the 3′ end of the important 153 bp interval there are two overlapping potential E2F binding sites present in opposite orientations (TTTGCCGG and ATTCCCGG; motifs described in ref. 22). These sites were present in the GL1::GUS reporter used to study GL1 expression in wild type and gl3-sst sim backgrounds.11 The presence of these potential E2F binding sites in the precise region defined to be important for GL1 expression supports the possibility that GL1 expression is directly tied to the cell cycle. Future experiments to test this region directly for E2F binding and to assess its chromatin conformation during trichome development will aid in strengthening the connection between GL1 expression and the cell cycle.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5471
References
- 1.Schellmann S, Hulskamp M. Epidermal differentiation: trichomes in Arabidopsis as a model system. Int J Dev Biol. 2005;49:579–584. doi: 10.1387/ijdb.051983ss. [DOI] [PubMed] [Google Scholar]
- 2.Schiefelbein J. Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr Opin Plant Biol. 2003;6:74–78. doi: 10.1016/s136952660200002x. [DOI] [PubMed] [Google Scholar]
- 3.Szymanski DB, Lloyd AM, Marks MD. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis Trends. Plant Sci. 2000;5:214–219. doi: 10.1016/s1360-1385(00)01597-1. [DOI] [PubMed] [Google Scholar]
- 4.Schnittger A, Hulskamp M. Trichome morphogenesis: a cell cycle perspective. Philos Trans R Soc Lond B Biol Sci. 2002;357:823–826. doi: 10.1098/rstb.2002.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- 6.Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 9.Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development. 2003;130:5885–5894. doi: 10.1242/dev.00812. [DOI] [PubMed] [Google Scholar]
- 10.Hulskamp M, Misra S, Jurgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-x. [DOI] [PubMed] [Google Scholar]
- 11.Marks MD, Gilding E, Wenger JP. Genetic interaction between glabra3-shapeshifter and siamese in Arabidopsis thaliana converts trichome precursors into cells with meristematic activity. Plant J. 2007;52:352–361. doi: 10.1111/j.1365-313X.2007.03243.x. [DOI] [PubMed] [Google Scholar]
- 12.Esch JJ, Chen MA, Hillestad M, Marks MD. Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 2004;40:860–869. doi: 10.1111/j.1365-313X.2004.02259.x. [DOI] [PubMed] [Google Scholar]
- 13.Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hulskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002;21:5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker JD, Oppenheimer DG, Concienne J, Larkin JC. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development. 2000;127:3931–3940. doi: 10.1242/dev.127.18.3931. [DOI] [PubMed] [Google Scholar]
- 15.Churchman ML, Brown ML, Kato N, Kirik V, Hulskamp M, Inze D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG, Gwin T, Churchman J, Larkin JC. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell. 2006;18:3145–3157. doi: 10.1105/tpc.106.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inze D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 17.Schnittger A, Schobinger U, Bouyer D, Weinl C, Stierhof YD, Hulskamp M. Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc Natl Acad Sci USA. 2002;99:6410–6415. doi: 10.1073/pnas.092657299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szymanski DB, Jilk RA, Pollock SM, Marks MD. Control of GL2 expression in Arabidopsis leaves and trichomes. Development. 1998;125:1161–1171. doi: 10.1242/dev.125.7.1161. [DOI] [PubMed] [Google Scholar]
- 19.Larkin JC, Oppenheimer DG, Pollock S, Marks MD. Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell. 1993;5:1739–1748. doi: 10.1105/tpc.5.12.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagami H, Sekine M, Murakami H, Shinmyo A. Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J. 1999;18:243–252. doi: 10.1046/j.1365-313x.1999.00449.x. [DOI] [PubMed] [Google Scholar]
- 21.Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A. Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell. 2002;14:1847–1857. doi: 10.1105/tpc.002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inze D, De Veylder L. Genome-wide identification of potential plant E2F target genes. Plant Physiol. 2005;139:316–328. doi: 10.1104/pp.105.066290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen WH. The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci. 2002;7:505–511. doi: 10.1016/s1360-1385(02)02351-8. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez-Parra E, Gutierrez C. E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 2007;144:105–120. doi: 10.1104/pp.106.094979. [DOI] [PMC free article] [PubMed] [Google Scholar]