Abstract
In plants, flowering is a critical developmental transition orchestrated by four regulatory pathways. Distinct alleles encoding mutant forms of the Arabidopsis potential calcium sensor CML24 cause alterations in flowering time. CML24 can act as a switch in the response to day length perception; loss-of-function cml24 mutants are late flowering under long days, whereas apparent gain of CML24 function results in early flowering. CML24 function is required for proper CONSTANS (CO) expression; components upstream of CO in the photoperiod pathway are largely unaffected in the cml24 mutants. In conjunction with CML23, a related calmodulin-like protein, CML24 also inhibits FLOWERING LOCUS C (FLC) expression and therefore impacts the autonomous regulatory pathway of the transition to flowering. Nitric oxide (NO) levels are elevated in cml23/cml24 double mutants and are largely responsible for FLC transcript accumulation. Therefore, CML23 and CML24 are potential calcium sensors that have partially overlapping function that may act to transduce calcium signals to regulate NO accumulation. In turn, NO levels influence the transition to flowering through both the photoperiod and autonomous regulatory pathways.
Key Words: calcium, nitric oxide, calmodulin, cell signaling, flowering
Introduction
The switch from leaf to flower production is critically important for plants. Many plants flower in response to seasonal day length. For example, 14 or more hours of light accelerate floral induction in Arabidopsis. This photoperiod pathway involves circadian regulation of CONSTANS (CO) transcription1,2 and the coincident perception of light that is necessary for CO protein stabilization.3 The autonomous pathway inhibits premature flowering through FLOWERING LOCUS C (FLC).4,5 FLC and other autonomous pathway genes also affect the circadian clock indicating crosstalk between the photoperiod and autonomous pathways.6
The photoperiod and autonomous pathways are also both affected by nitric oxide (NO).7 Mutants that overproduce NO are late flowering and have reduced CO and enhanced FLC expression; whereas the loss of function of a gene called NO ASSOCIATED 1 (AtNOA1) (previously called NO SYNTHASE 1, AtNOS1) results in early flowering, elevated CO expression, and reduced FLC expression.7,8 The mechanism that regulates NO accumulation during flowering is unclear.
Cytosolic calcium (Ca2+) levels oscillate with a 24-hour rhythm in plants.9 The amplitude and kinetics of Ca2+ fluctuations reflect light intensity and day-length onditions.10 Thus, Ca2+ oscillations have the potential to act as intracellular signals to integrate biological clock output and environmental stimuli and to regulate flowering transition. However, evidence for a functional role of the Ca2+ oscillations in plants has not yet been obtained.9
In many organisms, calcium (Ca2+) signals are sensed by calmodulin (CaM). Arabidopsis has a large family of CAM and CAM-like (CML) genes; the physiological functions of these potential Ca2+ sensors remain largely unknown.11,12 Arabidopsis with a silenced CML24 flower later than wild type when grown under long-day photo-periods.13 Here we report that CML24 and its close paralog CML23 regulate the transition to flowering by affecting CO and FLC expression and NO accumulation.
Results
CML23 and CML24 share sequence similarity and developmental expression patterns.
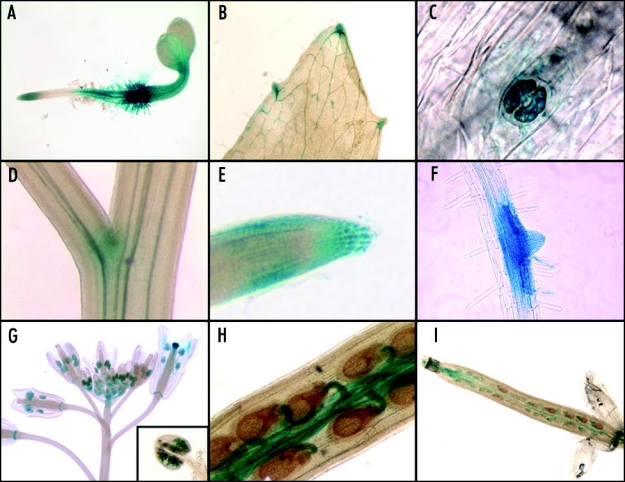
CML23 and CML24 likely arose by segmental duplication and encode proteins that share 78% sequence identity11 (Fig. 1). To gain insight into whether the paralogous genes may share overlapping function, we sought to compare expression characteristics of CML23 with that previously determined for CML24.13 RT-PCR indicates that CML23- and CML24-specific mRNAs accumulate in all major organs, including rosette and cauline leaves, roots, flowers and immature siliques13 (additional data not shown). CML24 expression levels are on average 14-fold greater than that of CML23, based on Genevestigator14 analysis. Approximately two kb of sequences found upstream of the CML23 translational start site were recombined with the GUS reporter gene, and six independent transgenics were generated and found to share similar GUS expression patterns (data not shown). CML23:GUS expression is detected throughout the hypocotyl, and diffusely in most central portions of the cotyledons, and the proximal and most distal root regions of four-day-old light-grown seedlings (Fig. 2A). CML23:GUS is expressed at hydathodes (Fig. 2B), guard cells (Fig. 2C) and inflorescence branch points (Fig. 2D). Epidermal, columellar and border-like cells of the root have CML23:GUS staining (Fig. 2E), in addition to emerging lateral roots and some root hairs (Fig. 2F). In developing flowers, CML23:GUS activity is in anthers and pollen (Fig. 2G and inset), and stigma of mature flowers (Fig. 2G). CML23:GUS expression is high in silique funiculi (Fig. 2H) and the abscission zone (Fig. 2G and I). Aspects of expression patterns detected with CML24:GUS transgenes13 are highly similar to that of CML23:GUS transgenics shown in Figure 2. For example, expression in stomata, hydathodes and silique abscission zones are indistinguishable between the two genes (Fig. 2).13 However, although CML23:GUS and CML24:GUS are both expressed at branching points, CML23:GUS staining is seen most prominently in the adaxial region of the branch (Fig. 2D), whereas CML24:GUS stains more uniformly and includes the abaxial branch region and portions of the primary inflorescence.13 Root tip expression is also distinct for the two genes. Whereas CML23:GUS expression is more restricted to the outer epidermal regions and the extreme root tip (Fig. 2E), CML24:GUS expression is absent from the distal tip but is in the meristematic and elongation zones.13 These analyses indicate that CML23 and CML24 are expressed in highly similar, yet non-identical patterns.
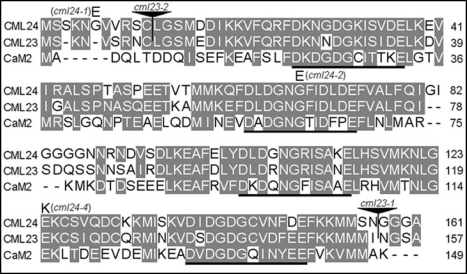
Figure 1.
CML23 and CML24 are calmodulin-like proteins. Amino acid sequence alignment of Arabidopsis CML24 (At5g37770), CML23 (At1g66400), and one of the calmodulins, CaM2 (At2g41110). Shaded amino acids indicate sequence similarity, underlines delineate EF-hand Ca2+ binding loops, letters above the CML24 sequence indicate amino acid substitutions in the cml24-1, cml24-2, and cml24-4 mutants, the solid triangles indicate the two independent insertion sites of the T-DNA insertions in CML23. Amino acid numbers are at right.
Figure 2.
CML23:GUS is expressed in many organs and throughout development. CML23:GUS is expressed in (A) four-day-old seedlings, (B) cauline leaves, (C) guard cells, (D) branch junctions on the inflorescence stem, (E) root tip, (F) sites of lateral root formation, (G) flower organs, (H) funiculi and (I) siliques.
Identification and molecular characterization of cml23 and cml24 mutants.
To probe the functions of the CML23 and CML24 genes, we isolated mutants. Tilling was used to identify point mutations in CML24.15 cml24-1 and cml24-2 are substitutions of glutamates (E) for glycines (G) at positions 6 and 67, respectively; cml24-4 has lysine (K) instead of E at amino acid 124 (Fig. 1). Two CML23 T-DNA insertion alleles were identified from the Salk collection.16 Sequencing from the left border verified the insertion sites (Fig. 1). CML23 transcripts are undetectable from either mutant using RT-PCR (data not shown), suggesting that cml23-1 and cml23-2 are null alleles.
To evaluate the potential consequences of the three point mutations on CML24 accumulation, we used anti-CML24 antibodies to detect the protein in extracts from wild type (Col-0) and cml24 mutants. SDS-PAGE followed by immunoblotting reveals that all the mutants accumulate CML24 protein (Fig. 3). Although Figure 3 suggests that there may be subtle differences in CML24 accumulation among the mutants, additional independent immunoblotting analyses indicate that these apparent quantitative differences are not consistently seen (data not shown). These results indicate that the cml24-1, cml24-2 and cml24-4 amino acid substitutions do not significantly affect overall protein accumulation.
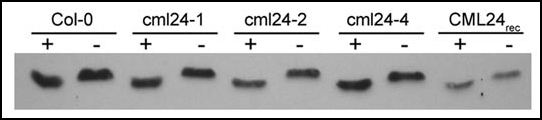
Figure 3.
Mutant CML24 proteins show a Ca2+-dependent mobility shift. Wild type (Col-0) and mutant CML24 proteins (cml24-1, cml24-2 and cml24-4) were analyzed along side purified CML24 produced heterologously in E. coli (CML24rec). Protein samples contained either 5 mM CaCl2 (+) or 5 mM EGTA (−) and were separated by 13% SDS-PAGE. The proteins were transferred to nitrocellulose membrane and probed with anti-CML24 antibody. The faster migration of wild-type, mutant, and recombinant CML24 in the presence of Ca2+ than in the presence of EGTA indicates that mutations in cml24-1, cml24-2 and cml24-4 do not completely impair Ca2+ binding and consequent conformational changes.
To begin to elucidate whether CML24 protein function is affected by the cml24-1, cml24-2 and cml24-4 amino acid substitutions, we assessed the ability of the mutant proteins to undergo Ca2+-dependent conformational changes. CML24, like CaMs, displays a Ca2+-dependent mobility shift in SDS-Page.13 Wild-type CML24, mutant CML24 proteins, and protein produced heterologously in E. coli run faster in the presence of Ca2+ than in the presence of EGTA (Fig. 3). Thus, these mutant CML24 proteins can bind Ca2+ and undergo conformational changes as a consequence. However, loss of a single Ca2+-binding site or modest reductions in Ca2+ binding affinities may not be detectable with this assay. For example, the amino acid substitution in cml24-2 is predicted to reduce Ca2+ affinity of the second EF hand because of the loss of the highly conserved G at position 6 in the Ca2+-binding loop. Future experiments with more sensitive Ca2+ affinity assays will reveal whether the mutations cause subtle alterations in CML24 biochemical activity and/or structural conformations. In addition, experiments are underway to test the possibility that the mutations affect the ability of CML24 to interact with target proteins.
The photoperiod pathway is altered in cml24 mutants.
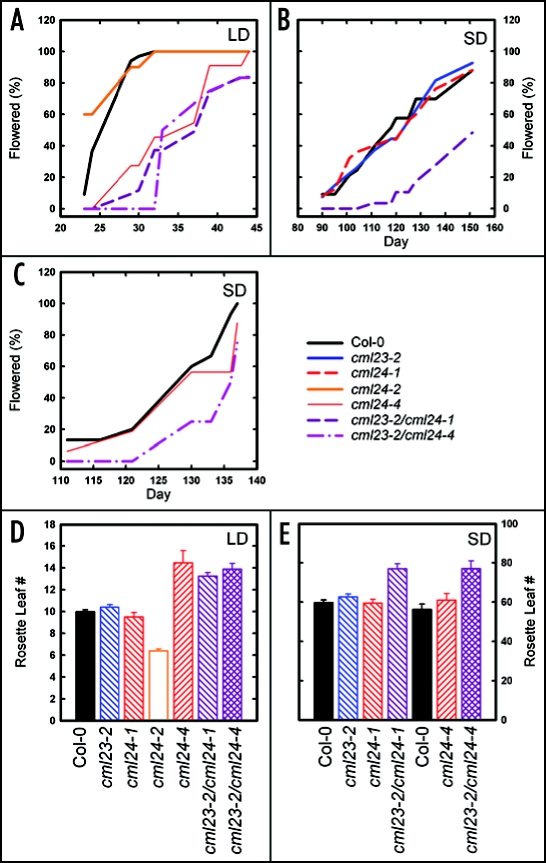
The cml24-4 mutant flowers later than wild type in long-day photoperiods of 16-hour (Fig. 4A) and 24-hour (data not shown) light and generates a greater number rosette leaves than wild type prior to flowering (Fig. 4D). Altered rosette leaf number at flowering is an indicator that flowering time differences are the consequence of defective regulation of floral induction and not simply altered growth rate. cml24-4 flowering is comparable to wild type when grown in short-day photoperiods (Fig. 4C and E), subjected to an extended cold period (data not shown), or treated with gibberellin (GA) (data not shown). These three results indicate, respectively, that the autonomous, vernalization and GA flowering regulatory pathways are intact.17 cml24-4 is thus specifically defective in the photoperiod pathway. The coincidence of a photoperiod-specific defect in both cml24-4 and CML24-silenced plants,13 for which little or no CML24 protein is detectable,13 suggests that the E124K mutation in cml24-4 causes at least a partial loss of CML24 function.
Figure 4.
CML23 and CML24 are required for appropriate timing of the transition to flowering. Wild type (Col-0) and mutants were grown in16-hour photoperiods (A) or short-day eight-hour (B) and seven-hour (C) photoperiods and the percentages of plants flowering over time were recorded. Rosette leaf numbers at flowering onset from (A–C) were recorded for Col-0 and mutants grown under (D) long-day and (E) short-day photoperiods. Values are means ± SEM (n = 6 to 33).
Conversely, cml24-2 flowers early (Fig. 4A) and produces fewer rosette leaves prior to flowering (Fig. 4D) in long-day photoperiods. cml24-2 flowering time resembles that of wild type in short days (data not shown). The opposing effect on flowering time of the cml24-2 and cml24-4 mutations suggests that the cml24-2 G67E mutation may result in a constitutively active CML24. Consistent with a possible gain of function, cml24-2 is a dominant mutation; plants heterozygous for the mutation also flower early under long-day conditions (data not shown). Together these data indicate that CML24 may act as a regulatory switch, stimulating flowering when activated and inhibiting flowering when inactive.
CML23 contributes to flowering regulation under long days.
Flowering time and rosette leaf number at flowering are not significantly affected in cml23-1, cml23-2 or cml24-1 under long or short photoperiods (Fig. 4B, D and E, and data not shown). However, when the cml23-2 and cml24-1 mutations are combined in double mutants, the transition to flowering in long days is delayed (Fig. 4A and D). These data indicate that CML23 and CML24 have overlapping roles in photoperiod regulation of flowering, that CML23 is dispensable in the presence of a wild-type CML24, and that cml24-1 is likely a weak allele that manifests a late-flowering phenotype only when CML23 is also defective.
Double mutants of cml23 and cml24 are defective in the autonomous pathway.
The double mutants also flower later than wild type under short photoperiods, revealing a role for the related Ca2+ sensors in the autonomous pathway. Relative to wild type, cml23-2/cm24-1 and cml23-2/cml24-4 flower later (Fig. 4B and C) and produce a greater number of leaves (Fig. 4E) under short-days of seven or eight hours of light. These double mutants respond to both vernalization and GA (data not shown), indicating that the CML23 and CML24 functions are likely limited to the photoperiod and autonomous pathways regulating flowering.
CML23 and CML24 act upstream of CO and FLC.
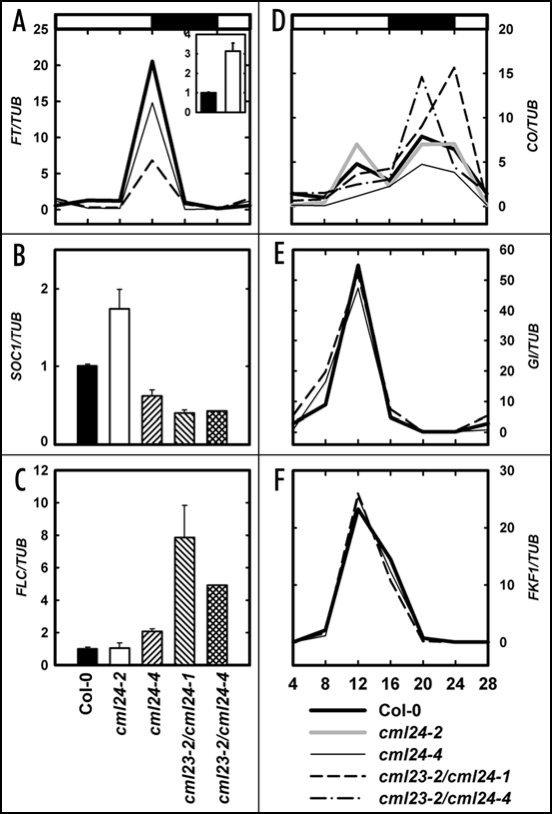
To begin to dissect where CML23 and CML24 act in the photoperiod and autonomous pathways, we analyzed expression of genes implicated in flowering regulation. FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) integrate regulatory information from both the photoperiod and autonomous pathways and promote flowering.17,18 The peak of FT transcript accumulation that occurs before the start of the dark transition is reduced in two late-flowering mutants analyzed, cml24-4 and cml23-2/cml24-1 (Fig. 5A). SOC1 expression is reduced in all the late-flowering mutants (Fig. 5B). In contrast, both FT and SOC1 expression levels are elevated in the early-flowering cml24-2 mutant (Fig. 5A and B). This altered gene expression in the cml mutants suggests that CML23 and CML24 act upstream of these pathway integrators.
Figure 5.
Regulation of flowering time gene expression in cml mutants. Wild type and cml mutants were grown for two weeks in 16-hour photoperiods and harvested at four-hour intervals over a 24-hour period (A, D, E and F) or 11 hours after dawn (A–C). Quantitative RT-PCR was performed to detect abundances of (A) FT, (B) SOC1, (C) FLC, (D) CO, (E) GI, (F) FKF1 transcripts relative to TUB4 (encoding tubulin) transcripts. In (A), Col-0 represented by the solid bar; cml24-2 is the open bar. The light and dark periods for (A) and (D–F) are represented by the open bars and filled bar, respectively, above (A and D). Values are means ± SEM (n = 3 to 5).
FT and SOC1 expression levels are dependent upon CO protein activity.19–21 CO activity is regulated through coincident signals from the circadian clock and photoreceptors, which convey when the photoperiod is appropriate for flowering.1,2 The late-flowering mutants cml24-4, cml23-2/cml24-1 and cml23-2/cml24-4 have reduced CO transcript accumulation during the day (Fig. 5D), the time when CO mRNA accumulation can lead to stable and thereby functional protein product.3 cml24-4 has the strongest effect, with a loss of 76% of the peak CO transcripts found in wild type at 12 hours after dawn (Fig. 5D). CO expression levels in cml23-2/cml24-1 and cml23-2/cml24-4 are reduced by 24% and 48%, respectively (Fig. 5D). In contrast, the early flowering mutant, cml24-2, has elevated daytime CO expression (Fig. 5D). CO, FT, and SOC1 expression levels are comparable to wild type in cml24-1 (data not shown). Taken together, these data indicate that CML24 is required for, and CML23 contributes to, the regulation of CO transcript accumulation during the day. The defect in daytime CO expression in the mutants may account, at least in part, for alterations in FT and SOC1 transcript levels and delayed flowering in long-day photoperiods.
Genes that act upstream of CO and the circadian clock appear largely unaffected by mutations in CML23 or CML24. Transcripts from GIGANTEA (GI) and FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1), which are required for proper CO expression,1,22,23 accumulate with nearly identical diurnal kinetics in wild type, cml24-4, and cml23-2/cml24-1 (Fig. 5E and F). However, GI transcripts do appear to begin to accumulate slightly earlier in the mutants (Fig. 5E). Leaf movements of the double mutants (cml23-2/cml24-1 and cml23-2/cml24-4) reveal a possible circadian period lengthening (data not shown); however, the effect is likely too modest to account for the late flowering behavior. In addition, circadian CO expression continues in long-day entrained cml23-2 and cml23-2/cml24-4 placed in constant light conditions (data not shown). CML23 and CML24, therefore, are unlikely to play major roles in the regulation of the circadian clock or GI or FKF1 expression; however these paralogous Ca2+ sensors may act either downstream or in parallel with these photoperiod pathway components and at a regulatory step upstream of CO.
CO expression in wild type reaches a second peak during the night (Fig. 5D); the mechanism of this regulation is not well defined. cml24-4 has reduced whereas cml23-2/cml24-1 and cml23-2/cml24-4 have higher CO expression than wild type during the night (Fig. 5D). CO expression is unaffected in cml23-2 mutants (data not shown); therefore, CML23 and CML24 have functional overlap in the darkness-specific repression of CO. The consequence of aberrant nighttime CO expression is, as yet, unclear because CO protein is unstable in the dark.3
Delayed flowering of double cml23/cml24, but not single, mutants in short-day photoperiods (Fig. 4B, C and E) indicate that CML23 and CML24 have overlapping functions regulating the autonomous pathway. FLC transcript levels are elevated several fold in cml23-2/cml24-1 and cml23-2/cml24-4 (Fig. 5C) and altered expression is detectable in the shoot apex (data not shown) where FLC is normally expressed.24 These results are consistent with the mutants flowering late in short days (Fig. 4B, C, and E). The single mutants, cml23-2, cml24-1 and cml24-4, show no or only modest increases in FLC transcript accumulation (Fig. 5C and data not shown). The coordinate mis-regulation of both CO and FLC expression in the double mutants may explain why FT and SOC expression levels are more strongly affected in, for example, cml23-2/cml24-1 than in cml24-4 which is only modestly affected in FLC transcript levels. Although mutation of CML24 can be sufficient to affect photoperiod-regulated flowering time, the autonomous pathway is affected only when both CML23 and CML24 are impaired. We conclude that CML23 and CML24 act upstream of FLC to inhibit expression and thus promote flowering.
cml23 and cml24 mutants accumulate nitric oxide.
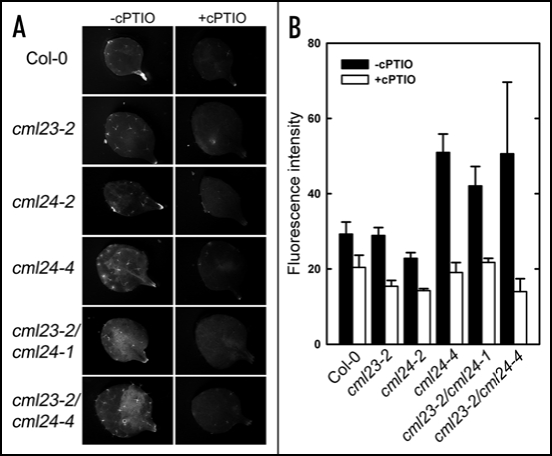
NO has been shown to affect both the autonomous and photoperiod pathways by affecting FLC, GI and CO expression.7 Because NO and the CML23/CML24 proteins were found to both act in the regulation of FLC and CO, we wanted to determine whether the effects of cml23/cml24 mutations were the consequence of altered NO accumulation regulation. Therefore, we compared NO levels in the cml23 and cml24 mutants with wild-type plants. NO accumulation was monitored with an NO-sensitive fluorescent dye, 4-amino-5-methyl-amino-2′,7′-di-fluorofluorescein (DAF-FM DA) (Fig. 6). Plants were grown in 16 hours of light per day and under axenic conditions to avoid possible effects of plant-associated microbes on NO levels. Comparable fluorescence is detected in wild-type, cml23-2 and cml24-2 leaves (Fig. 6A and B). In contrast, the late-flowering mutants, cml24-4, cml23-2/cml24-1 and cml23-2/cml24-4, have 1.5- to 1.8-fold elevated fluorescence (Fig. 6A and B). These elevated DAF-FM fluorescence levels are comparable in magnitude to that seen in the late flowering NO over producer nox1.7 The elevated fluorescence in the late-flowering cml mutants is most likely due to higher NO accumulation because incubation with cPTIO, an NO scavenger, significantly reduces the DAF-FM fluorescence (Fig. 6A and B). Although DAF dyes can fluorescence in response to compounds likely distinct from NO itself, non-NO specific fluorescence is not reduced by cPTIO.25 Taken together these data indicate that loss of CML23 and CML24 results in elevated NO in leaves.
Figure 6.
CML23 and CML24 regulate NO accumulation in leaves. (A) Rosette leaves of two-week-old Col-0 and cml mutants were treated with or without 0.4 mM cPTIO (NO scavenger) for four hours and then stained with DAF-FM DA (NO sensitive fluorescence dye). Fluorescence was detected with 490–495 nm excitation and 515 nm emission. (B) Average fluorescence intensity levels from the rosette leaves corresponding to the treatments in (A) were quantified using Image J (NIH). Values are means ± SEM (n = 6).
Aberrant NO accumulation is responsible for altered FLC expression in the cml Mutants.
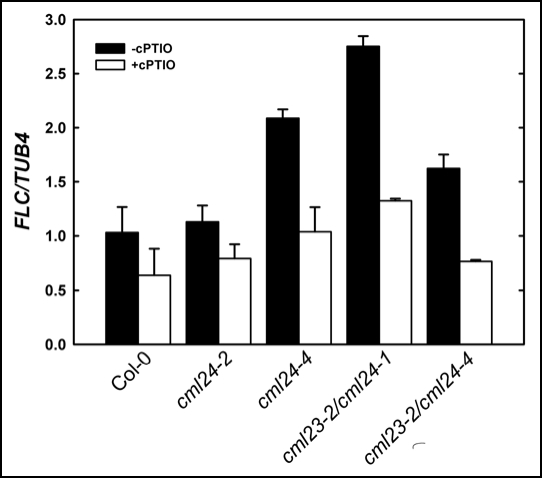
To investigate whether the elevated NO levels in the cml23/cml24 mutants may be responsible for alterations in gene expression, we quantified FLC transcript abundance in wild-type and mutant plants grown under a 16-hour photoperiod, and treated with cPTIO to reduce NO levels. A 4-hour cPTIO treatment has no significant effect on FLC transcript levels in wild type or cml24-2, but strongly reduces FLC expression in the late-flowering cml24-4, cml23-2/cml24-1 and cml23-2/cml24-4 mutants (Fig. 7). These results suggest that although NO may not be required for basal FLC expression, elevated NO is essential for the enhanced FLC expression in the late flowering cml24-4, cml23-2/cml24-1 and cml23-2/cml24-4 mutants.
Figure 7.
Altered NO levels affect FLC transcript levels. Quantitative RT-PCR of FLC levels relative to TUB4 were determined in 2-week-old plants grown under long-day (16-hr) photoperiods and treated with or without 0.4 mM cPTIO for 4 hours. Values are means ± SEM (n = 3 to 4).
Discussion
CML23 and CML24: Potential Ca2+ sensors with non-identical but overlapping function.
The Arabidopsis genome contains seven CaM-encoding genes and an additional 50 genes encoding CaM-like proteins.11 The physiological functions of these potential Ca2+ sensors remain largely unknown, despite the central role Ca2+ is thought to play as a second messenger. Comparative molecular modeling of CML24 based on the paramecium CaM crystal structure reveals that despite its sequence divergence from CaM, CML24 likely has an overall structure that closely resembles CaM with two globular domains separated by a linker helix.26 Like CaM, CML24 also binds Ca2+ and undergoes Ca2+-dependent conformational changes exposing hydrophobic patches.13 These characteristics indicate that CML24 is likely to be a CaM-like Ca2+ sensor. However, the sequence divergence of CML24 from CaMs argues that CML24 has functions that are distinct from those of CaM. This work provides genetic evidence for this idea because the CaMs and other CMLs do not compensate for mutations in CML24 and the related paralog CML23. Therefore, distinct plant Ca2+-binding proteins likely have specialized cellular and physiological functions, as has been documented for animal CaM-like proteins.27
Tilling28 is a powerful technique to identify point mutations in genes of interest. Although we identified no point mutations that resulted in stop codons, we were able to isolate three distinct cml24 mutants described here (Fig. 1).
Using Ca2+-dependent SDS-PAGE mobility shifts as an assay, we find that the mutant CML24 isoforms can all bind Ca2+ and change conformation (Fig. 3). However, because this assay may not reveal more subtle changes in Ca2+ binding, such as loss of Ca2+ binding by only one of the four predicted Ca2+-binding sites or a decrease in Ca2+ affinity, we are undertaking further biophysical studies to fully ascertain the affect of the three mutations on the Ca2+-binding abilities of the mutant CML24 proteins. It is also possible that the amino acid substitutions have no effect on Ca2+-binding but instead alter the ability of the CML24 proteins to interact with and/or regulate the activity of potential proteins. These possibilities are under current investigation.
Phenotypic analysis reveals that the late-flowering phenotype of cml24-4 resembles that of cml24 gene-silenced lines (Fig. 4).13 Therefore the cml24-4 mutation likely results in at least a partial loss of function. cml24-1 results in detectable nonwild-type phenotypes only when combined with a mutant cml23 gene (Figs. 4–6). Therefore, cml24-1 is likely a weaker allele than cml24-4. Distinctions between the cml23-2/cml24-1 and cml23-2/cml24-4 mutant phenotypes may be explained by the difference in allele strength of the cml24-1 and cml24-4 mutations. Alternatively, distinct amino acid substitutions may differentially affect CML24 interactions with different targets. For example, distinct CaM mutants differentially interact with and regulate distinct target proteins and thus result in different downstream effects.29,30 To shed light on the distinct functional consequences of the different cml24 point mutations, we are identifying and isolating proteins that interact with CML24 and may be downstream targets. To date, null mutations of CML24 have not been identified. Although gene silencing approaches have the potential to be useful in characterizing full loss of CML24 function, we found CML24 RNAi lines to be unstable (data not shown), and our concern about possible cross reactivity silencing related genes and potentially complicating interpretation has led us to focus our attention on the Tilling lines.
The cml24-2 mutation results in a dominant phenotype that is opposite in character to cml24-4 and CML24 gene silenced lines (Figs. 4 and 5)13 and therefore may be a gain of function mutant. Tilling thus enabled the identification of an allelic series of mutations that reveal that CML24 may act as a switch to control flowering time. Reduction or loss of CML24 function delays whereas gain of CML24 function accelerates the transition to flowering under long-day conditions (Fig. 4).
CML23 is the closest relative to CML24, sharing 78% amino acid sequence identity with CML24 (Fig. 1).11 The paralogs likely derived from a segmental chromosome duplication revealed in part by their common placement adjacent to the paralogous CAM genes, CAM1 and CAM4, which encode identical proteins.11 CML23 and CML24 share highly similar, yet nonidentical, development patterns of expression (Fig. 2);13 CML23 expression levels, however, are less than 10 percent that of CML24. Although phenotypic consequences of cml23 mutations have not yet been detected, combination of a cml23 null allele with a weak cml24 mutation (cml24-1) results in late-flowering and over accumulation of NO (Figs. 4–7). Therefore, CML23 and CML24 share overlapping, but not identical, physiological functions.
Roles for potential Ca2+ sensors in regulation of the transition to flowering.
Ca2+ levels fluctuate in response to many stimuli, including stimuli that affect flowering time, such as circadian rhythms,10 light31,32 and temperature.33–35 However, whether these Ca2+ fluctuations are responsible for regulating aspects of the flowering transition is unknown. The findings that the potential Ca2+ sensors CML23 and CML24 play roles in regulating the transition to flowering (Fig. 4) suggest the possibility that these proteins may integrate Ca2+ second messengers generated in response to the endogenous biological clock and exogenous environmental signals into the flowering regulatory pathways.
Mutations in CML24 alone affect the photoperiod pathway (Fig. 4A and D), whereas deletion of CML23 has no significant effect on flowering (Fig. 4B, D and E). Combining mutations in the two genes impacts both the photoperiod and the autonomous pathways (Fig. 4A–E). Thus, loss of CML23 reveals additional functions for CML24 in the autonomous pathway. However, the loss of CML23, when combined with CML24 mutations, also affects the photoperiod pathway. That is, the photoperiod defects in cml23/cml24 double mutants are not identical to defects of the cml24-4 single mutants. However, because aberrant autonomous pathway regulation can impact photoperiod pathway circadian rhythms,6 it is possible that CML23 affects photoperiod pathway regulation indirectly through its function in the autonomous pathway.
Roles for potential Ca2+ sensors in regulation of nitric oxide accumulation.
Plants, like animals, use NO as a signaling molecule. NO has been implicated in diverse physiological processes.36 However, the major pathway(s) by which NO is generated is not clear. Two enzymatic pathways have been implicated in NO production in Arabidopsis, nitrate reductase (NR) and nitric oxide associated 1 (AtNOA1, previously called NOS1).8,37,38 AtNOA1-dependent NO production is required for proper transition to flowering.7 NO levels correlate with FLC expression levels and the timing of the transition to flowering.7 Scavenging NO can reduce FLC transcript accumulation in the late-flowering cml mutants (Fig. 7) consistent with the idea that elevated NO is necessary to promote elevated FLC expression. How NO levels are regulated in response to flowering pathway signals or environmental stress is not known. Elevated NO levels in cml23/cml24 mutants (Fig. 6) indicate that CML23 and CML24 have roles in inhibiting NO accumulation in plants. CML23:GUS (Fig. 2) and CML24:GUS13 expression in roots is high in the distal portion of the transition zone from which NO production is detected.38,39 One possibility is that the potential Ca2+ sensors directly affect the activities of NR or AtNOA1; alternatively, CML23 and CML24 may affect nonenzymatic NO production and/or NO scavenging.40,41 CaM directly regulates animal NO synthase.42 If CaM has a similar stimulatory activity in plants, CaM-related proteins, such as CML23 and CML24, may act as CaM competitive antagonists to block NO synthesis. Reciprocal regulation by two soybean CaM isoforms of mammalian NOS can occur in vitro.43
In summary, we have demonstrated overlapping roles for the Arabidopsis CML23 and CML24 potential Ca2+ sensor proteins in regulating the transition to flowering and NO accumulation. CML23 and CML24 thus may function to integrate signaling information from cytosolic Ca2+ to the production and/or degradation of cellular NO to regulate flowering in Arabidopsis.
Methods
Plant material and growth conditions.
Insertional mutants were obtained as segregating T3 populations from the Arabidopsis Biological Resource Center;16 Tilling mutants were obtained as homozygous and heterozygous populations from the Arabidopsis Tilling Project.28 Mutants were back crossed at least twice and all Arabidopsis used in this study are of the Col-0 ecotype. Plants were cultivated and flowering time assays were conducted as (described in ref. 13).
Generation and staining of the CML23:GUS transgenics.
To generate the CML23:GUS plasmid the forward primer, 5′GGGG CTGCAGCTGTCTTAATATTTGGTTGTACTAGA3′, and the reverse primer, 5′GGGGGATCCTTTTTTAGAGAGAAATAGAA GA 3′, were used to amplify 1906 basepairs of sequences upstream of the CML23 translational start site. The amplicon was then directionally ligated into the PstI/BamHI sites of the pCB301 binary vector44 engineered to include the β-GLUCURONIDASE (GUS) coding sequence and NOS termination sequence. The underlined bases introduce the restriction enzyme sites into the PCR product.
Agrobacterium transformation of Arabidopsis was performed using the floral dip method.45 CML23:GUS transgenics were submerged in a 100 mM Na-phosphate buffer, pH 7.0 staining solution containing 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid with cyanide.46 Staining was visualized and photographed using the Zeiss axioscope (Carl Zeiss, Germany) and Leica MZFL III stereoscope (Switzerland).
Identification of homozygous cml23 and cml24 single and double mutants.
To identify homozygous cml23 insertional mutants, PCR using CML23-specific primers (5′GGACATGTCGA AGAACGTTTCGAGAAACTG3′ and 5′ CTGGCGCGCCAGAG AGCCATTAAAGAAGCAAC3′) and T-DNA specific primers (5′ A CTTGATTTGGGTGATGGTTCACGTAG3′ and 5′ GCAATAAT GGTTTCTGACGTATGTGCT3′) was performed. Products generated were sequenced by Lone Star Sequencing (Houston, TX) to determine exact insertion sites and extent of sequence deletions. Point mutations generated and identified by Tilling were detected by differential restriction enzyme digestion of gene-specific PCR products. The following primer sets were used to identify mutants: cml24-1(G6E), 5′ATAAAGATGCCACCAGCTCACGCAATCTC3′ and 5′TTGGCGCGCCTCAAGCACCACCACCATTACTCATC ATCTTCTT3′, cml24-2 (G67E), 5′AACAGCATCACCAGAAGA AACAGT3′ and 5′GAAAAGCGCGACGAATTCGTCCAGATCT ATGGAT3′; and cml24-4 (E124K), 5′AAACAATTCGATCTAGA CGGTAACGGATTC3′ and 5′TACGAATCATCACCGTCGACT AAT3′. CML24 PCR products from segregating progeny of heterozygous cml24-1 plants were digested with XmnI; similarly, cml24-2 PCR product was digested with BamHI, and cml24-4 PCR product was cut with HphI to identify homozygous mutants. Double mutants were generated by crossing cml23 T-DNA insertion mutants with cml24 point mutants. Homozygous mutants were identified from the F2 or later generations using the PCR- and restriction digest-based analyses described above.
Immunoblotting.
Total plant protein was extracted from roots using a lysis buffer (4% [w/v] SDS, 20% [v/v] glycerol, 120 mM Tris, pH 6.8) as (described in Ref. 47). Recombinant CML24 was purified by binding to phenyl-sepharose, washing with 1 mM CaCl2, and eluting with 1 mM EGTA. Protein concentration was determined using the Pierce BCA kit. Twenty micrograms of total plant protein from wild type (Col-0) or mutant plants (cml23-2/cml24-1, cml24-2, and cml24-4) or 1 µg of recombinant CML24 protein were separated in a 13% (w/v) SDS-polyacrylamide gel. Protein extracts were was boiled in the presence of 5 mM EGTA or 5 mM CaCl2 before subjection to SDS-PAGE. The proteins were transferred to nitrocellulose membrane in transfer buffer (25 mM Tris, 192 mM Glycine, 1 mM CaCl2, 20% [v/v] methanol, pH 8.3).48,49 Blots were baked six hours at 65°C in a vacuum oven to enhance CML24 protein fixation onto the membrane.47 The membrane was incubated with 2.4 µg/mL of anti-CML24 primary antibody47 followed by Pierce horseradish peroxidase-conjugate goat anti-rabbit secondary antibody (Rockford, IL). The antibody solutions were diluted in 150 mM NaCl, 10 mM Tris-HCl, pH 7.5, 0.1% (v/v) Tween 20, and 1% (w/v) nonfat milk. The protein bands were detected using the Pierce SuperSignal West Pico Chemluminescent Kit (Rockford, IL).
Quantitative real-time RT-PCR (Q RT-PCR).
Q RT-PCR was performed as described in reference 13. To analyze gene expression, total RNA was extracted using TRI REAGENT (Molecular Research Center, Cincinnati, OH) according to the manufacturer′s instructions. The first strand cDNA was synthesized from 1 µg of RNA following DNase treatment and reverse transcription using DNase (Roche Diagnostics) and SuperScript III Reverse Transcriptase (Invitrogen), respectively. The CO, FLC, FKF1, FT, GI and SOC1 transcripts were assessed by quantitative real-time RT-PCR in an ABI PRISM 7000 (Applied Biosystems) using Thermocycler ABI SYBR Green PCR Master Mix (Applied Biosystems). The difference in cycle number where product amplification resulted in a fixed threshold amount of fluorescence was determined by the following equation:
ΔCTsample = ΔCTTUB4−ΔCTgene of interest.
One sample was chosen as a calibrator and ΔΔCT was determined for each sample according to the equation:
ΔΔCTsample=ΔCTsample−ΔCTcalibrator.
Relative RNA levels were calculated using inverse log2, 2^ (ΔΔCTsample). The primer sequences for Q RT-PCR were: CO, 5′AT ATGGCTCCTCAGGGACTC3′ and 5′GGGTCAGGTTGTTGC TCTAC3′; FLC, 5′AAGAAGAGAACCAGGTTTTG3′ and 5′GAAGATTGTCGGAGATTTGT3′; FKF1, 5′CGTTAGAGG TTGGGATGTTC3′ and 5′CGAGGATCTCTGTACTGTAG3′; FT, 5′TGATATCCCTGCTACAACTG3′ and 5′TCGCGAGTGT TGAAGTTCTG3′; GI, 5′GATTGCTGCTCCTGAAATCC3′ and 5′GATGCACTTGCGAGAATCAC3′; SOC1, 5′CAAGCAGAC AAGTGACTTTC3′ and 5′GCCTCAGATAACGATCTATG3′; and TUB4, 5′CTGTTTCCGTACCCTCAAGC3′ and 5′AGGGAAAC GAAGACAGCAAG3′.
Nitric oxide staining, quantitation and manipulation.
To monitor nitric oxide levels in vivo, the first and second true leaves were detached from two-week-old plants and incubated in MES buffer (10 mM MES (2-(N-morpholino) ethanesulphonic acid), 10 mM KCl, 0.1 mM CaCl2, pH 5.8) for two hours. After addition of 10 µM 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM DA, Molecular Probes) for 45 minutes, fluorescence was analyzed with excitation at 490–495 nm and emission at 515 nm and visualized using a MZ FLIII (Leica, Switzerland) fluorescence stereomicroscope. All the images had the same exposure time. To quantify the DAF-FM DA fluorescence, images were analyzed by ImageJ 1.36b (Wayner Rasband, National Institute of Health, USA). The NO scavenger, 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO, Sigma-Aldrich) (400 µM in 0.4%DMSO, 0.1% Tween20), was directly sprayed on leaves of two-week-old plants; the control leaves were sprayed with solvent (0.4% DMSO, 0.1% Tween20). Leaves were harvested after four hours.
Acknowledgements
This material is based upon work supported by the National Science Foundation under grant No. IBN0321532 and IBN0313432 and by the Department of Energy Grant number DE-FG02-03ER15394. We are grateful to Patrice A. Salome and Rob McClung for conducting assays of circadian leaf movement, the Arabidopsis Tilling Project for providing the CML24 point mutants, the Salk Institute Genomic Analysis Laboratory for providing the CML23 insertion mutants, the Arabidopsis Biological Resource Center for seed stocks, and Richard Gomer, Bonnie Bartel, Mike Gustin and James McNew for critical comments on the manuscript.
Footnotes
Both authors contributed equally to this work.
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4695
References
- 1.Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. Constans mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 2.Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- 3.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 4.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheldon CS, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salathia N, Davis SJ, Lynn JR, Michaels SD, Amasino RM, Millar AJ. FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biol. 2006;6:10. doi: 10.1186/1471-2229-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, Fiorani F, Jackson RB, Crawford NM, Pei ZM. Nitric oxide represses the Arabidopsis floral transition. Science. 2004;305:1968–1971. doi: 10.1126/science.1098837. [DOI] [PubMed] [Google Scholar]
- 8.Crawford NM, Galli M, Tischner R, Heimer YM, Okamoto M, Mack A. Response to Zemojtel et al: Plant nitric oxide synthase: Back to square one. Trends Plant Sci. 2006;11:526–527. [Google Scholar]
- 9.Dodd AN, Love J, Webb AAR. The plant clock shows its metal: Circadian regulation of cytosolic free Ca2+ Trends Plant Sci. 2005;10:15–21. doi: 10.1016/j.tplants.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Love J, Dodd AN, Webb AAR. Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell. 2004;16:956–966. doi: 10.1105/tpc.020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCormack E, Braam J. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 2003;159:585–598. doi: 10.1046/j.1469-8137.2003.00845.x. [DOI] [PubMed] [Google Scholar]
- 12.McCormack E, Tsai YC, Braam J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005;10:383–389. doi: 10.1016/j.tplants.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Delk NA, Johnson KA, Chowdhury NI, Braam J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to ABA, day length and ion stress. Plant Physiol. 2005;139:240–253. doi: 10.1104/pp.105.062612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. Genevestigator: Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCabe PF, Valentine TA, Forsbert LS, Pennell RI. Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. Plant Cell. 1997;9:2225–2241. doi: 10.1105/tpc.9.12.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 17.Simpson GG, Dean C. Arabidopsis, the rosetta stone of flowering time? Science. 2002;296:285–289. doi: 10.1126/science.296.5566.285. [DOI] [PubMed] [Google Scholar]
- 18.Mouradov A, Cremer F, Coupland G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell. 2002;14:S111–S130. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kardailsky I, Shukla VK, Anh JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 20.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 22.Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 23.Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, Coupland G. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell. 2005;17:2225–2270. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 25.Planchet E, Kaiser WM. Nitric oxide production in plants. Plant Signal Behav. 2006;1:46–51. doi: 10.4161/psb.1.2.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan AR, Johnson KA, Braam J, James MNG. Comparative modeling of the three-dimensional structure of the calmodulin-related TCH2 protein from Arabidopsis. Proteins. 1997;27:144–153. doi: 10.1002/(sici)1097-0134(199701)27:1<144::aid-prot14>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 27.Ikura M, Ames JB. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: Two ways to promote multifunctionality. Proc Natl Acad Sci USA. 2006;103:1159–1164. doi: 10.1073/pnas.0508640103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCallum CM, Comai L, Greene EA, Henikoff S. Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 2000;123:439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okano H, Cyert MS, Ohya Y. Importance of phenylalanine residues of yeast calmodulin for target binding and activation. J Biol Chem. 1998;273:26375–26382. doi: 10.1074/jbc.273.41.26375. [DOI] [PubMed] [Google Scholar]
- 30.Ohya Y, Botstein D. Diverse essential functions revealed by complementing yeast calmodulin mutants. Science. 1994;263:963–966. doi: 10.1126/science.8310294. [DOI] [PubMed] [Google Scholar]
- 31.Ries S, Widders I, Baughan R. Light Affects the Flux of Ca2+ from excised tomato leaves within four seconds. J Exp Bot. 1995;46:1867–1876. [Google Scholar]
- 32.Long JC, Jenkins GI. Involvement of plasma membrane redox activity and calcium homeostasis in the UV-B and UV-A /bluelight induction of gene expression in Arabidopsis. Plant Cell. 1998;10:2077–2086. doi: 10.1105/tpc.10.12.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plieth C, Hansen UP, Knight H, Knight MR. Temperature sensing by plants: The primary characteristics of signal perception and calcium response. Plant J. 1999;18:491–497. doi: 10.1046/j.1365-313x.1999.00471.x. [DOI] [PubMed] [Google Scholar]
- 34.Wood NT, Allan AC, Haley A, Viry-Moussaid M, Trewavas AJ. The characterization of differential calcium signalling in tobacco guard cells. Plant J. 2000;24:335–344. doi: 10.1046/j.1365-313x.2000.00881.x. [DOI] [PubMed] [Google Scholar]
- 35.Knight H, Knight MR. Imaging spatial and cellular characteristics of low temperature calcium signature after cold acclimation in Arabidopsis. J Exp Botany. 2000;51:1679–1686. doi: 10.1093/jexbot/51.351.1679. [DOI] [PubMed] [Google Scholar]
- 36.Beligni MV, Lamattina L. Nitric oxide in plants: The history is just beginning. Plant Cell Environ. 2001;24:267–278. [Google Scholar]
- 37.Crawford NM. Mechanisms for nitric oxide synthesis in plants. J Exp Bot. 2006;57:471–478. doi: 10.1093/jxb/erj050. [DOI] [PubMed] [Google Scholar]
- 38.Guo FQ, Okamoto M, Crawford NM. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science. 2003;302:100–103. doi: 10.1126/science.1086770. [DOI] [PubMed] [Google Scholar]
- 39.Illes P, Schlicht M, Pavlovkin J, Lichtscheidl I, Baluska F, Ovecka M. Aluminium toxicity in plants: Internalization of aluminium into cells of the transition zone in Arabidopsis root apices related to changes in plasma membrane potential, endosomal behavior, and nitric oxide production. J Exp Bot. 2006;57:4201–4213. doi: 10.1093/jxb/erl197. [DOI] [PubMed] [Google Scholar]
- 40.Bethke PC, Badger MR, Jones RL. Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell. 2004;16:332–341. doi: 10.1105/tpc.017822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kundu S, Trent JTI, Hargrove MS. Plants, humans and hemoglobins. Trends Plant Sci. 2003;8:387–393. doi: 10.1016/S1360-1385(03)00163-8. [DOI] [PubMed] [Google Scholar]
- 42.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: Structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho MJ, Vaghy PL, Kondo R, Lee SH, Davis JP, Rehl R, Heo WD, Johnson JD. Reciprocal regulation of mammalian nitric oxide synthase and calcineurin by plant calmodulin isoforms. Biochemistry. 1998;37:15593–15597. doi: 10.1021/bi981497g. [DOI] [PubMed] [Google Scholar]
- 44.Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. A mini binary vector series for plant transformations. Plant Mol Biol. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- 45.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 46.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3906. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sistrunk ML, Antosiewicz DM, Purugganan MM, Braam J. Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell. 1994;6:1553–1565. doi: 10.1105/tpc.6.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 49.McKeon TA, Lyman ML. Calcium ion improves electrophoretic transfer of calmodulin and other small proteins. Anal Biochem. 1991;193:125–130. doi: 10.1016/0003-2697(91)90051-t. [DOI] [PubMed] [Google Scholar]