Abstract
The wheat ALMT1 gene encodes an aluminum (Al)-activated malate transport protein which confers Al-resistance. We investigated the membrane topology of this plasma-membrane localized protein with immunocytochemical techniques. Several green fluorescent protein (GFP)-fused and histidine (His)-tagged chimeras of ALMT1 were prepared based on a computer-predicted secondary structure and transiently expressed in cultured mammalian cells. Antibodies raised to polypeptide epitopes of ALMT1 were used in conjunction with the antibody to the His-tags to determine the topology of ALMT1. This study shows that the ALMT1 protein contains six transmembrane domains with the amino and carboxyl termini located on the extracellular side of the plasma membrane.
Key Words: ALMT1, aluminum resistance, immunofluorescent staining, malate transporter, topology, Triticum aestivum
Introduction
Aluminum ions present in acid soils are toxic to plants, and poor plant growth can often be directly correlated with the soluble Al concentrations in soil solutions.1,2
The efflux of organic acids from roots plays an important role in the Al resistance exhibited by some plant species.3,4 For instance, Al-resistant wheat plants release malate anions from their root cells which chelate the harmful Al cations in the apoplasm.5–7 Malate efflux is controlled by the ALMT1 gene which encodes an Al-activated malate transport protein localized to the plasma membrane of root cells.8–12 Heterologus expression of ALMT1 confers an Al-activated malate efflux in Xenopus laevis oocytes, rice, barley and cultured tobacco cells, which, in tobacco cells and barley plants, is associated with enhanced Al resistance.8,13
Elucidation of the secondary structure of a membrane protein can result in a better understanding of protein function by defining regions of the protein that are intra- or extra-cellular and by relating these regions to protein function. For instance, an understanding of which regions of the protein are exposed to the apolplasm or symplasm of a cell may provide clues to how protein function is regulated by metabolic processes or influenced by extracellular signals. The topology of wheat ALMT1 is likely to be similar in both plant and animal membranes, since the protein is functional when heterologously expressed in Xenopus oocytes as well as plant cells.8 In a previous report, human embryonic kidney 293 cells were successfully used for the topological analysis of the Na+/K+ translocating AtHKT1 from Arabidopsis.14 In this study, we used the same experimental system to transiently express a range of ALMT1 protein tagged with GFP or histidine residues and used immunocytochemical techniques to determine the secondary structure of ALMT1. This paper describes the first topological analysis of ALMT1.
Material and Methods
Plasmid constructs.
The mammalian expression vector pcDNA3.1 (Invitrogen, Carlsbad, USA) was used to prepare constructs designed to fuse ALMT1 with (1) GFP at the carboxyl (C)-terminus (ALMT1::GFP); (2) six histidine residues (His-tag) at the amino (N)-terminus (His::ALMT1) or (3) a His-tag at the C-terminus (ALMT1::His) (see RESULTS). The ALMT1::GFP fragment derived from pTH-ALMT1::GFP9,15 was inserted into the XbaI and NotI sites of pcDNA3.1 to construct pcDNA3.1-ALMT1::GFP. To prepare pcDNA3.1-His::ALMT1 and pcDNA3.1-ALMT1::His, the coding sequence of ALMT1-1 was amplified by PCR using the primers 5′-GGTCTAGAATGATGGATATTGATCACGGCAGAGAG -3′ (sense) and 5′-G GGATATCTTACAAAATAACCACGTCAGGCAAAGG-3′ (antisense) for His::ALMT1, or the primers 5′-GGTCTAGAATGGATATTGATCACGGCAGAGAG-3′ (sense) and 5′-GGGATATCTTAGTGGTGGTGGTGGTGGTGCAAAATAACCACGTCAGGCAAAGG-3′ (antisense) for ALMT1::His. The amplified fragments of ALMT1 containing coding sequence for the His-tag (double underlined) were digested at the Xba I and Not I sites (underlined) and inserted into the Xba I and Not I sites of pcDNA3.1.
To generate His epitopes in the putative internal loops of ALMT1 by site-directed mutagenesis, the ALMT1 cDNA was subcloned into the pGEM-T easy vector (Promega, Madison, USA), and the entire loop 2 (F76-L79) and a part of loop 3 (S103-R108), loop 4 (E130-Q135) and loop 5 (P159-K164) were replaced with the DNA fragment coding six histidine residues (L2h, L3h, L4h, L5h, see Results and Table 1), using the QuikChange® mutagenesis kit (Stratagene, La Jolla, USA). The individual mutants were verified by sequencing, and the His-tagged ALMT1 fragments were inserted into the NotI sites of pcDNA3.1.
Table 1.
His-tagged ALMT1 proteins created by replacement of several amino acids with His residues
Generation of antipeptide antibodies.
Rabbit polyclonal antisera (Labfrontier, Suwon, Korea) were raised against the polypeptide epitopes (NTp, L1p, L3p, L4p, L6p, CTHp; see Results and Table 2) localized at N-terminal, C-terminal half and putative internal loops (1, 3, 4, 6) of ALMT1.
Table 2.
Amino acid sequences of peptide epitopes of ALMT1 used to immunize rabbits for raising antibodies
| Name of Epitope Site | Amino Acid Sequences at Epitope Site1 | ||
| Epitope | Amino Acid Positions | Amino Acid Sequences | |
| NTp | N-terminal | 1–13 | MDIDHGRESDGEM |
| L1p | Loop 1 | 41–52 | KVGGAAREDPRR |
| L3p | Loop 3 | 103–108 | SKGLNR |
| L4p | Loop 4 | 127–135 | ELAERCGDQ |
| L6p | Loop 6 | 187–196 | EELIQLAHQR |
| CTHp | C-terminal half | 253–264 | NNFGGKDFPQMH |
The sites of these peptides in ALMT1 proteins are shown in Figure 1B.
Cell culture, transfection and immunocytochemical analyses.
Human embryonic cells (293T) and C3H10T1/2 mouse fibroblast (10T1/2 cells) were cultured in Dulbecco's Modified Eagle Medium supplemented with 10% heat-inactivated fetal bovine serum and gentamicin (50 µg/ml). Cells plated onto culture chambers (Nunc, Naperville, USA) were transfected with plasmid DNA using GeneJuice transfection reagent (Novagen, Madison, USA) for 48 h. The transfection frequency was approximately 5% to 30%, judging from the ratio of the ALMT1::GFP transformants among cells. The cells were then fixed using 4% paraformaldehyde and incubated in phosphate-buffered saline (PBS) for 15 min without or with 0.5% Triton X-100 to permeabilize the cell membrane. Cells were blocked with 3% bovine serum albumin (BSA) in PBS for 1 h and then incubated for 1 h at room temperature with primary antibodies diluted in PBS containing 3% BSA (1:250–1:1,000 dilution). The antibodies consisted of either mouse anti-His antibody (Covance, Princeton, USA) or the various rabbit antibodies raised against ALMT1 polypeptide epitopes (see above). Cells were rinsed three times in PBS and then incubated with the secondary antibodies (1:800 dilution) of either goat anti-mouse IgG conjugated with alexa fluor 488 (AF488) or goat anti-rabbit IgG conjugated with alexa fluor 594 (AF594) (Molecular Probes, Eugene, USA) in PBS containing 3% BSA for 1 h. Cells were washed three times in PBS, mounted on glass slides with Perma Fluor (Immunon, Pittsburg, USA), and the fluorescence form AF488, AF594 and GFP autofluorescences was detected using the filter sets B, G and B, respectively, with a Fluorescence microscope (CKX41N-31FL/PHP, Olympus, Tokyo, Japan).
Results
Computer-predicted domains.
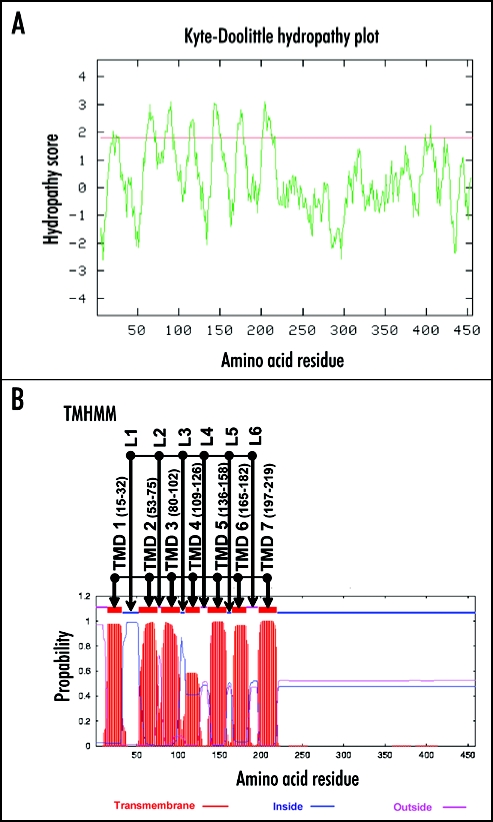
The Kyte-Doolittle algorithm provides hydrophobicity scores (gcat.davidson.edu/rakarnik/kytedoolittle.htm),16 and the TMHMM hidden Markov model predicts transmembrane helices in protein sequences (v2.0; www.cbs.dtu.dk/services/TMHMM-2.0/).17 Both algorithms predicted that the N-terminal half of ALMT1 (M1-A219) to be hydrophobic and to possess seven transmembrane domains (TMD 1 to TMD 7), with the first and fourth domains being less certain than the others (Fig. 1). To test this model, we systematically investigated the orientation of the N- and C-termini and the six hydrophilic loops (L1 to L6) with immunocytochemical techniques.
Figure 1.
Transmembrane domain prediction models applied to ALMT1. (A) shows the hydrophobicity of ALMT1 predicted by the Kyte-Doolittle algorithm. Peaks with hydropathy scores greater than 1.8 (red line) indicate likely transmembrane regions. (B) shows the prediction of transmembrane domains of ALMT1 by the TMHMM algorithm. Seven transmembrane domains (TMD 1 to TMD 7, with the amino acid positions of the ends of the domains) with the relative probability on the Y axis and six loop regions (L1 to L6) are shown.
The amino- and carboxy-termini of ALMT1 are located extracellularly.
Preliminary experiments indicated that tobacco cells (intact cells, protoplasts) were unsuitable for the topological analyses using the immunocytochemical techniques described below because of non-specific binding of the antipeptide antibodies raised against ALMT1 on both ALMT1 transformants and non transformants of tobacco cells (data not shown). By contrast, the mammalian cells (293T, 10T1/2) expressing ALMT1 did not suffer from this problem.
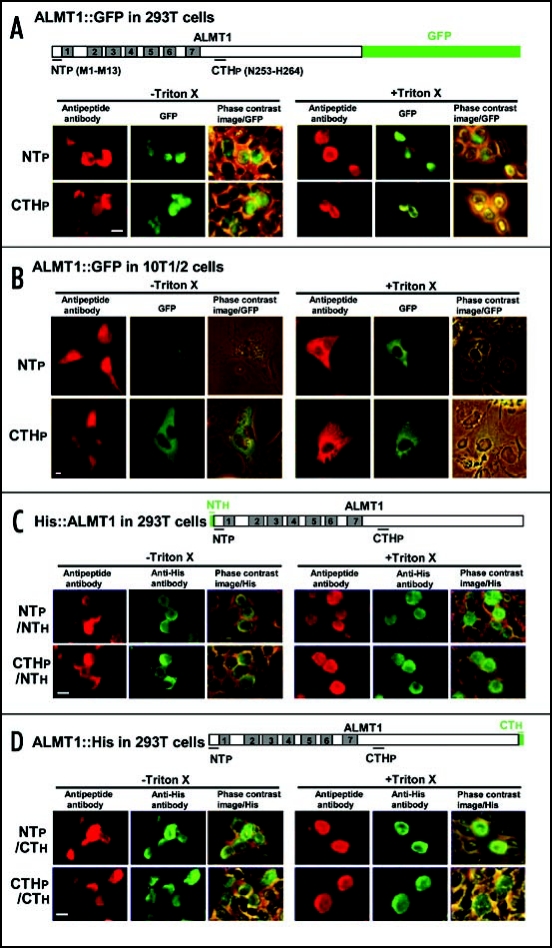
The mammalian cells were firstly transformed with a construct encoding an ALMT1::GFP fusion protein, and the transformants were identified by the green fluorescence of GFP (Fig. 2A and B). GFP fluorescence was used only to identify cells expressing the ALMT1::GFP fusion proteins and not for topological analyses, because GFP fluorescence is autofluorescence and could be detected directly without any help of fluorochrome-conjugated secondary antibodies in the ALMT1::GFP transformants regardless of detergent treatment and regardless whether the protein was located intra- or extra-cellularly. Furthermore, we found that the intensity of the GFP autofluorescence was reduced strongly after the cells were fixed with paraformaldehyde prior to the immunocytochemical reactions (see Materials and Methods; data not shown). Indeed, although both GFP and AF594 fluorescence were detected in the same cells (Fig. 2A and B), the localization patterns for these fluorescence were slightly different at times (see below). These results suggest that the fixative seems to distort the secondary structure of the GFP protein and to affect its capacity to fluoresce, whereas the fluorochrome-conjugated secondary antibodies applied after the fixative treatment are not affected by the fixative.
Figure 2.
The orientation of the N- and C-termini of ALMT1 expressed in mammalian cells (293T, 10T1/2). Cells transiently expressing the ALMT1 construct fused with GFP at the C-terminus (A and B), tagged with a His epitope at the N-terminus [NTh, (C)] or tagged with a His epitope at the C-terminus [CTh, (D)] were identified with green GFP fluorescence and the green AF488 fluorescence obtained with anti-His antibody, respectively, in the presence or absence of Triton X-100. These cells were simultaneously tested for the accessibility of the polypeptide epitopes located at the N-terminus (NTp) and C-terminal half region (CTHp) to their respective anti-peptide antibodies (detected by red AF594 fluorescence). Representative images are shown here. Left panels, AF594 fluorescence; Center panels, GFP fluorescence or AF488 fluorescence; Right panels, the phase contrast image overlaid with the GFP or AF488 fluorescence. Bar = 20 µm.
To determine the topology, the ALMT1::GFP transformants were firstly identified by GFP fluorescence, and the orientation of the N- and C-terminal ends of ALMT1 was determined by raising rabbit antipeptide antibodies against a polypeptide epitope on the N-terminus (NTp) of the protein and against an epitope on the C-terminal half (CTHp) of the protein near the start of the long hydrophilic tail region (C-terminal half region, amino acid positions 220–459) predicted by the computer model. Transfected cells were challenged with the antibodies in the absence or presence of Triton X-100 and subsequently with anti-rabbit IgG conjugated with AF594. Triton X-100 treatment permeabilized the cells and allowed the antipeptide antibodies to bind with any epitopes accessible from the intracellular side of the membrane. Binding of the antipeptide antibody to their epitopes yields red AF594 fluorescence under a fluorescence microscope. It was clearly seen that the red AF594 fluorescence generated by the NTp and CTHp epitopes were both detected in the transformed 293T cells (Fig. 2A) and 10T1/2 cells (Fig. 2B) with or without treatment with Triton X-100. These results indicate that the antipeptide antibodies recognized their epitopes on the extracellular side of the plasma membrane.
To confirm the orientation of NTp and CTHp, we expressed the ALMT1 proteins tagged with His epitopes at the N-terminus (His::ALMT1) or C-terminus (ALMT1::His) in 293T cells. The cells were treated simultaneously with mouse anti-His antibodies and the rabbit antipeptide (NTp, CTHp) antibodies, and then subsequently with secondary antibodies of anti-mouse IgG conjugated with AF488 and anti-rabbit IgG conjugated with AF594. Once again, red AF594 and green AF488 fluorescence was detected together in the same cells, regardless of the Triton X-100 treatment (Fig. 2C and D), indicating that the epitopes on the N and C termini which were recognized by the anti-His and the antipeptide antibodies were extracellular. Interestingly, the fluorescence patterns generated by the AF594 and AF488 fluorophors were similar with or without Triton X-100 treatment (Fig. 2C and D), whereas the fluorescence from AF594 and GFP did not always show the same pattern (Fig. 2A and B), probably due to the distortion of GFP structure by paraformaldehyde as described above.
Orientation of putative loop regions.
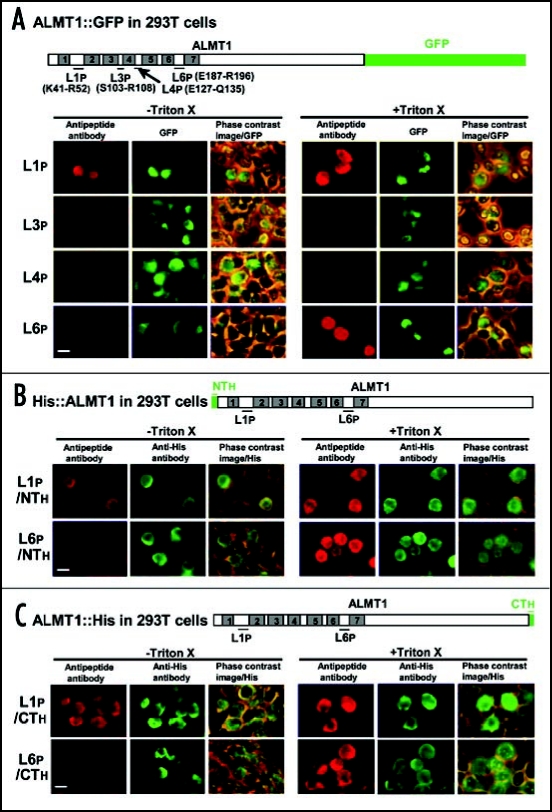
To determine the orientation of the putative loop regions, 293T cells expressing ALMT1::GFP, His::ALMT1 or ALMT1::His were challenged with rabbit antipeptide antibodies raised against the polypeptide epitopes at four loop regions (L1p, L3p, L4p, L6p) (Fig. 3). The rabbit anti-L1p antibody was detected with anti-rabbit IgG (red AF594 fluorescence) in the cells expressing ALMT1::GFP (green GFP fluorescence), whether cells were permeabilized or not with Triton X-100 (Fig. 3A), indicating that the L1 region was extracellular. This result was confirmed by similar experiments with cells expressing His::ALMT1 (Fig. 3B) and ALMT1::His (Fig. 3C) (detected by green AF488) which were also detected regardless of the Triton X-100 treatment. For the anti-L6 antibody, red AF594 signal was detected in the cells expressing ALMT1::GFP, His::ALMT1 and ALMT1::His only after membrane permeabilization with Triton X-100 (Fig. 3A–C), suggesting that the L6 region faces the cytosol. Antibodies raised against L3p and L4p were not detected in the cells expressing ALMT1::GFP regardless of the Triton X-100 treatment (Fig. 3A).
Figure 3.
The orientation of putative loop regions (L1, L3, L4, L6) of ALMT1 expressed in 293T cells. Cells transiently expressing the ALMT1 construct fused with GFP at the C-terminus (A) (detected by GFP fluorescence) or tagged with His at the N-terminus (B) or C-terminus (C) (detected by AF488 fluorescence) were identified as described in Figure 2, in the presence or absence of Triton X-100. These cells were simultaneously tested for the accessibility of the polypeptide epitopes located at L1 (L1p, shown with the amino acid positions of the ends of the epitope) and L6 (L6p) to their respective anti-peptide antibodies (detected by red AF594 fluorescence). Left panels, AF594 fluorescence; Center panels, GFP fluorescence or AF488 fluorescence; Right panels, the phase contrast image overlaid with the GFP or AF488 fluorescence. Bar = 20 µm.
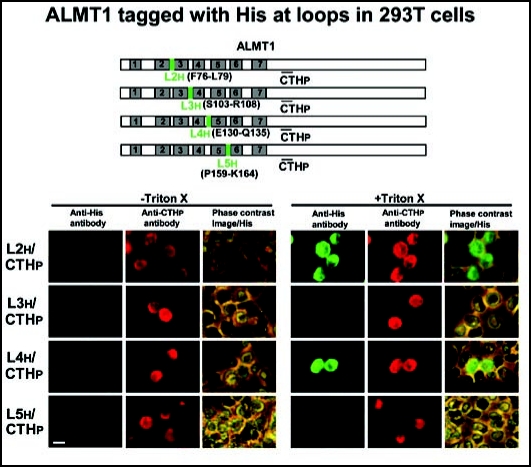
To confirm and extend these results using another method, we prepared constructs that tagged ALMT1 with His residues at L2 (L2h), L3 (L3h), L4 (L4h) and L5 (L5h), and expressed them in 293T cells (Fig. 4). L2h and L4h epitopes were recognized by the anti-His antibody (detected by green AF488) in the 293T cells expressing the ALMT1 CTHp epitope (detected by red AF594) only after the treatment with Triton X-100 (Fig. 4). The remaining epitopes, L3h and L5h, were not detected by the anti-His antibody with or without Triton X-100 treatment (Fig. 4).
Figure 4.
The orientation of putative loop regions (L2, L3, L4, L5) of ALMT1 expressed in 293T cells. Cells transiently expressing the ALMT1 construct tagged with His at L2 (L2h, shown with the amino acid positions of the replacement to His tag), L3 (L3h), L4 (L4h) and L5 (L5h) were identified with the AF594 fluorescence obtained with the antipeptide antibody against CTHp, in the presence or absence of Triton X-100. These cells were simultaneously tested for the accessibility of the anti-His antibody (detected by AF488 fluorescence). Left panels, AF488 fluorescence; Center panels, AF594 fluorescence; Right panels, the phase contrast image overlaid with AF488 fluorescence. Bar = 20 µm.
We conclude that the predicted three loops, L2, L4 and L6, are indeed present in ALMT1, and that they all face the cytosol. The status of L3 and L5 is less certain. They could either be localized in the membrane where they cannot interact with the antibodies or, as predicted from the computer model, they could be extracellular but unable to react sufficiently with the antibodies due to their relatively small size.
Discussion
We used a computer-generated model as a basis for determining the secondary structure of the novel malate transporter ALMT1. Our immunocytochemical analyses (Figs. 2–4) generated a model where both the N- and C-terminal ends of ALMT1 are extracellular (Fig. 5). Although our experimental evidence largely agrees with the computer model, the first transmembrane domain predicted by the TMHMM algorithm was, in fact, found not to span the membrane but to be orientated extracellularly. Putative loops L2, L4 and L6 face the cytosol while the localization of L3 and L5 remains equivocal since the antibodies did not recognize the peptides and/or His epitopes on these regions. It is possible that the L3 and L5 regions are embedded inside the membrane because the computer model (Fig. 1B) not only predicts these loops to be short (six residues) but the putative transmembrane domains 4 and 6 are also shorter than the other transmembrane domains (18 residues compared with 23 residues). In addition, there may be possible interactions between these loop regions and other regions of the same ALMT1 protein. Alternatively these loop regions might be involved in forming multimeric structures with other proteins which prevents the antibodies from binding to them.
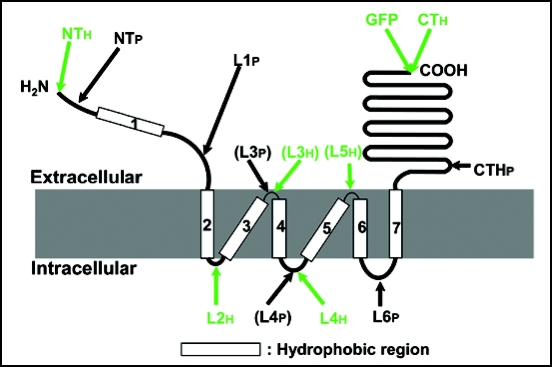
Figure 5.
Topological model of ALMT1. The model depicts the membrane topology of ALMT1 as determined by experimental evidence derived from the accessibility of antibodies to the epitopes (shown with arrows) when ALMT1 is expressed in mammalian cells (Figs. 2–4). The ALMT1 protein consists of hydrophobic N-terminal half and hydrophilic C-terminal half regions. The C-terminal half region (240 amino acids including CTHp) is extracellular. The N-terminal region, including the N-terminus, putative transmembrane domain 1 (TMD 1) and putative loop 1 (L1), is extracellular. Six TMDs follow (2 to 7) and three loops (L2, L4, L6) faced the cytsol. L4 was not detected by antipeptide antibody (shown in parentheses) but was detected with an anti-His antibody. However, both L3 and L5 were not detected even with the anti-His antibody (L3 was not detected by antipeptide antibody either and is shown in parentheses), suggesting that these loops are too small to be detected by the antibodies or could be embedded inside the membrane.
Taken together, we conclude that the ALMT1 protein has six transmembrane domains and three intracellular loops with both termini being extracellular.
To validate the conclusion, it may be necessary to determine whether the ALMT1 protein expressed in human 293T cells (or mouse 10T1/2 cells) confers Al-activated malate efflux. However, it was difficult to evaluate the ALMT1 function in this system, because the transformation frequency with the ALMT1 gene was low (approximately 5–30%) and the ALMT1 gene was only transiently expressed in 293T cells. To overcome this problem, we plan to generate 293T transformants stably expressing ALMT1. Alternatively, judging from our previous results that the ALMT1 protein expressed in Xenopus oocytes exhibits Al-activated malate efflux as found in cultured tobacco cells and barley roots,8,13 it is likely that functional ALMT1 protein is expressed in human 293T cells.
ALMT1 does not belong to any existing protein family of known function. One ALMT1 ortholog in Arabidopsis thaliana18 and two orthologs in Brassica napus19 also encode Al-activated malate transport proteins and possess similar hydropathy plots to ALMT1. Furthermore, several ALMT1 orthologs have been reported as putative ALMT1 transporters in rice (Oryza sativa, accession number. CAD40928 etc), rye (Secale cereale, ABA62397, ABA62398), maize (Zea mays, ABC86748), broccoli (Brassica oleracea, AAW81734) and barrel medic (Medicago truncatula, ABD32785, ABD32184). These ALMT proteins belong to a larger ALMT family, the members of which are largely uncharacterised.12 Nevertheless, hydrophobicity plots for all these putative proteins are similar, especially in the N-terminal half where the transmembrane domains occur (data not shown). Interestingly, the first transmembrane domain (TMD1) does not always exist in the orthologs, suggesting that this first hydrophobic region of ALMT1 may not be essential for the protein function.
The activation of organic anion (malate and citrate) channels by Al is an unusual response observed in Al-resistant plants7–9,18–21 and the mechanism of this activation remains unknown. A secondary structure of the ALMT1 protein provides the first step toward understanding how this protein functions and, in particular, how it is activated by Al. For instance, since it is now apparent that the relatively long and hydrophilic C-terminal region is orientated outside the cell, we can attempt mutagenesis to determine whether this region interacts with Al in the external solution to activate malate efflux.
Acknowledgements
We thank Yasuo Niwa (Univ. Shizuoka, Japan) for kindly providing the pTH2 vector carrying the GFP gene. This work was supported in part by Grant Program of The Sumitomo Foundation, Grant-in-Aid for Scientific Research (Nos. 17380049, 17078007, 18208008) and Special Educational Study on “Crop Improvement by Gene Analyses” from the Ministry of Education, Culture, Sports, Science and Technology of Japan, “2006 Research Grant for Encouragement of Students” and the COE program “Establishment of Plant Health Science” from Okayama University and Ohara Foundation for Agricultural Science.
Abbreviations
- Al
aluminum
- ALMT1
aluminum-activated malate transporter
- GFP
green fluorescent protein
- His
histidine
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4801
References
- 1.Kochian LV, Hoekenga OA, Piñeros MA. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Physiol Plant Mol Biol. 2004;55:459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]
- 2.Matsumoto H. Cell biology of aluminum toxicity and tolerance in higher plants. Int Rev Cytol. 2000;200:1–40. doi: 10.1016/s0074-7696(00)00001-2. [DOI] [PubMed] [Google Scholar]
- 3.Ryan PR, Delhaize E, Jones DL. Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:527–560. doi: 10.1146/annurev.arplant.52.1.527. [DOI] [PubMed] [Google Scholar]
- 4.Ma JF, Ryan PR, Delhaize E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001;6:273–278. doi: 10.1016/s1360-1385(01)01961-6. [DOI] [PubMed] [Google Scholar]
- 5.Delhaize E, Ryan PR, Randall PJ. Aluminum tolerance in wheat (Triticum aestivum L.). II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol. 1993;103:695–702. doi: 10.1104/pp.103.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan PR, Skerrett M, Flindlay G, Delhaize E, Tyerman SD. Aluminium activates an anion channel in the apical cells of wheat roots. Proc Natl Acad Sci USA. 1997;94:6547–6552. doi: 10.1073/pnas.94.12.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang WH, Ryan PR, Tyerman SD. Malate-permeable channels and cation channels activated by aluminum in the apical cells of wheat roots. Plant Physiol. 2001;125:1459–1472. doi: 10.1104/pp.125.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004;37:645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi M, Sasaki T, Sivaguru M, Yamamoto Y, Osawa H, Ahn SJ, Matsumoto H. Evidence for the plasma membrane localization of Al activated malate transporter (ALMT1) Plant Cell Physiol. 2005;46:812–816. doi: 10.1093/pcp/pci083. [DOI] [PubMed] [Google Scholar]
- 10.Raman H, Zhang K, Cakir M, Appels R, Garvin DF, Maron LG, Kochian LV, Moroni JS, Raman R, Imtiaz M, Drake-Brockman F, Waters I, Martin P, Sasaki T, Yamamoto Y, Matsumoto H, Hebb DM, Delhaize E, Ryan PR. Molecular characterization and maping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.) Genome. 2005;48:781–791. doi: 10.1139/g05-054. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki T, Ryan PR, Delhaize E, Hebb DM, Ogihara Y, Kawaura K, Noda K, Kojima T, Toyoda A, Matsumoto H, Yamamoto Y. Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant Cell Physiol. 2006;47:1343–1354. doi: 10.1093/pcp/pcl002. [DOI] [PubMed] [Google Scholar]
- 12.Delhaize E, Gruber BD, Ryan PR. The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Lett. 2007;581:2255–2262. doi: 10.1016/j.febslet.2007.03.057. [DOI] [PubMed] [Google Scholar]
- 13.Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H. Engineering high level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA. 2004;101:15249–15254. doi: 10.1073/pnas.0406258101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato Y, Sakaguchi M, Mori Y, Saito K, Nakamura T, Bakker EP, Sato Y, Goshima S, Uozumi N. Evidence in support of a four transmembrane-pore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc Natl Acad Sci USA. 2001;98:6488–6493. doi: 10.1073/pnas.101556598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu WI, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 16.Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 17.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 18.Hoekenga O, Maron LG, Pineros MA, Cancado GM, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, Matsumoto H, Yamamoto Y, Koyama H, Kochian LV. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:9734–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ligaba A, Katsuhara M, Ryan PR, Shibasaka M, Matsumoto H. The BnALMT1 and BnALMT2 genes from Brassica napus L. encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006;142:1294–1303. doi: 10.1104/pp.106.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piñeros MA, Kochian LV. A patch-clamp study on the physiology of aluminum toxicity and aluminum tolerance in maize: Identification and characterization of Al3+-induced anion channels. Plant Physiol. 2001;125:292–305. doi: 10.1104/pp.125.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kollmeier M, Dietrich P, Bauer CS, Horst WJ, Hedrich R. Aluminum activates a citrate-permeable anion channel in the aluminum-sensitive zone of the maize root apex: A comparison between an aluminum-sensitive and an aluminum-resistant cultivar. Plant Physiol. 2001;126:397–410. doi: 10.1104/pp.126.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]