Abstract
Adaxial/abaxial (dorsoventral) leaf polarity is a unique example of a developmental process in which early patterning decisions are determined by small regulatory RNAs. In maize, an abaxial gradient of the microRNA miR166 in the incipient leaf defines the localization of HD-ZIPIII transcripts that promote adaxial identity. Loss of leafbladeless1 (lbl1) function, necessary for the biogenesis of TAS3-derived trans-acting short-interfering RNAs (ta-siRNAs), leads to ectopic accumulation of miR166 throughout the initiating leaf. These findings indicate that the TAS3 ta-siRNA pathway specifies leaf polarity by spatially restricting miR166 to the abaxial side. Here, we briefly discuss possible mechanisms by which the TAS3 ta-siRNA pathway could regulate miR166 expression. Such regulatory mechanisms likely involve AUXIN RESPoNSE FACToRS and the phytohormone auxin. The convergence of small RNAs, and perhaps auxin, on key determinants of adaxial/abaxial organ polarity implicates them as candidate mobile signals that act at the plant apex to relay positional information from the meristem to the initiating leaf.
Key Words: microRNAs, trans-acting siRNAs, leaf polarity, maize
Patterning and outgrowth of leaves in higher plants depend on the specification of adaxial/abaxial (dorsoventral) polarity in the incipient leaf. Dorsoventrality is established through the polarized expression of class III homeodomain leucine zipper (HD-ZIPIII) transcription factors that specify adaxial/upper cell fate.1,2 Additional transcription factors, such as members of the AUXIN RESPONSE FACTOR (ARF) family, ARF3/ETTIN (ETT) and ARF4, act redundantly and in combination with KANADI proteins to promote abaxial fate.3
Interestingly, the adaxializing HD-ZIPIII genes, as well as the abaxial determinants ARF3/ETT and ARF4, are targets for RNAi-based regulation. Expression of HD-ZIPIII family members is regulated by the miR166 class of microRNAs, which direct HD-ZIPIII transcript cleavage.2,4 In maize, an abaxial gradient of miR166 within the incipient primordium restricts HD-ZIPIII expression to the adaxial surface, thereby establishing leaf polarity.2 ARF3/ETT and ARF4 are targets of 21-nt tasiR-ARFs, which are generated from TAS3 loci and belong to the recently discovered plant-specific class of small RNAs called trans-acting siRNAs (ta-siRNAs).5
The significance of HD-ZIPIII regulation by miR166 has been demonstrated through the characterization of dominant mutations in the HD-ZIPIII genes that disrupt the miR166 target site. Such mutations perturb miRNA-directed transcript cleavage, leading to ectopic, abaxial HD-ZIPIII expression and the formation of adaxialized leaves.1,2,6,7 In contrast, the contribution of tasiR-ARF-based regulation of ARF3/ETT and ARF4 to leaf polarity is not immediately evident. Arabidopsis mutants that block the biogenesis of ta-siRNAs develop accelerated juvenile-to-adult transition phenotypes rather than adaxial/abaxial polarity defects.5,8,9 Furthermore, the distribution of trichomes in such mutants, as well as in plants expressing a tasiR-ARF-insensitive allele of ARF3/ETT, is inconsistent with the predicted abaxializing phenotype.8,10,11
We have recently demonstrated a critical role for the TAS3 ta-siRNA pathway in organ polarity by cloning and characterizing leafbladeless1 (lbl1) from maize.12 Loss-of-function lbl1 mutations condition an abaxialized leaf phenotype and lead to loss of HD-ZIPIII expression, indicating that lbl1 is required, in addition to the HD-ZIPIII genes, to specify adaxial fate.12–14 lbl1 encodes an ortholog of the Arabidopsis SUPPRESSOR-OF-GENE-SILENCING3 (SGS3) protein, which is essential for ta-siRNA biogenesis.5,12
The TAS3 ta-siRNA pathway is conserved between Arabidopsis and maize, including the generation of tasiR-ARFs that direct the cleavage of ARF3 transcripts. Despite this conservation, the TAS3 ta-siRNA pathway has distinct roles in leaf polarity in both species. We showed that maize components of this pathway are expressed on the adaxial side of the newly initiated leaf, and restrict the expression domains of the abaxial factors arf3a and miR166 early in leaf development.12 The lack of obvious polarity defects in Arabidopsis TAS3 ta-siRNA pathway mutants may reflect a requirement for this pathway only during later stages of leaf development, to maintain organ polarity rather than establish it. In addition, the absence of polarity defects in Arabidopsis may reflect redundancy of the TAS3 ta-siRNA pathway with other polarity pathways, such as ASYMMETRIC LEAVES1/2 (AS1/2).15 We also envision that the phenotypic differences between maize and Arabidopsis are due to divergence in downstream components. The abaxial-promoting factor FILAMENTOUS FLOWER (FIL), for instance, is repressed by the Arabidopsis TAS3 ta-siRNA pathway, whereas members of the maize yabby gene family closely related to FIL are positively regulated by lbl1.14–16 Moreover, disruption of TAS3-derived ta-siRNA biogenesis in lbl1 leads to ectopic miR166 expression in the incipient and developing leaves.
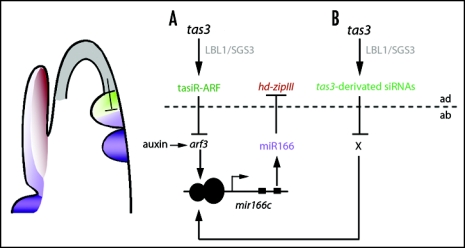
The maize genome includes at least nine mir166 loci, mir166a through mir166i, which generate identical mature miR166 molecules. We showed that primary transcripts for mir166c accumulate specifically in the lbl1 SAM and young leaf primordia, and expression levels for mir166i are increased in these mutant tissues. We therefore propose that the TAS3 ta-siRNA pathway defines the adaxial side of the leaf by restricting the expression domain of abaxial determinants, including mir166c and mir166i.12 We are currently exploring the mechanisms by which the TAS3 ta-siRNA pathway might control miR166 expression. We showed that in lbl1, ARF3 transcripts are misexpressed on the adaxial side of the initiating leaf. ARF proteins are transcription factors that mediate auxin-dependent gene regulation through binding to specific sequence motifs within promoters of auxin-regulated genes.17 miR166c contains a putative ARF binding site ∼800-bp upstream from the hairpin, suggesting the possibility that tasiR-ARFs control mir166c expression via ARF3 (Fig. 1A). However, ARF binding sites are not found in the mir166i promoter. This gene is either indirectly regulated by ARF3 or perhaps through unknown ta-siRNAs targets. Additional ta-siRNAs are produced from the maize TAS3 loci (Fig. 1B). Although targets for such ta-siRNAs remain elusive, it is conceivable that they may regulate the expression of mir166 family members as well.
Figure 1.
Models for adaxial/abaxial patterning via TAS3-derivated ta-siRNAs and miR166. (A) LBL1/SGS3 is required for the biogenesis of tasiR-ARFs from tas3 loci on the adaxial (ad) side of the incipient primordium that guide the cleavage of ARF3 transcripts. ARF3 may directly regulate the expression of specific mir166 family members, such as mir166c. (B) Additional TAS3-derivated siRNAs may guide the cleavage of novel targets (X) that directly or indirectly regulate the expression of mir166 family members. Mature miR166 accumulates on the abaxial (ab) side of the initiating leaf and confines the expression of the HD-ZIPIII family members to the adaxial surface. The opposing activities of TAS3-derivated ta-siRNAs and miR166 thus specifies polarity in the developing leaves.
Regulation of mir166 genes by ARF proteins is an attractive hypothesis because it invokes a contribution of auxin, a mobile signal, in creating the observed expression gradient of miR166 in the incipient leaf.2 However, the existence of auxin gradients18 in developing leaves and a direct contribution of auxin to adaxial/abaxial patterning remain to be established. Ta-siRNAs are generated through a unique branch of the RNAi machinery, which may limit the accumulation or efficacy of these small RNAs. In addition, because components of this distinct RNAi branch generate mobile siRNAs required for systemic silencing,19 we proposed that ta-siRNAs or biogenesis components might act non-cell-autonomously over a few cell layers. If so, the graded pattern of miR166 accumulation may also be generated by an inverse concentration gradient of TAS3-derivated ta-siRNAs formed across the initiating leaf, such that a precise balance between ta-siRNAs and miR166 activities specifies organ polarity.12
Adaxial/abaxial patterning is thought to depend on instructive, possibly non-cell autonomous signals that relay positional information inherent within the shoot apical meristem to polarized expression patterns in the incipient primordium.20 The recognition that TAS3-derived ta-siRNAs and likely auxin regulate ARF3/ETT and ARF4 suggests candidate positional signals that converge on the regulation of miR166 to establish organ polarity. As the involvement of the TAS3 ta-siRNA pathway in leaf polarity differs between plant lineages, it is possible that the contribution of small RNAs and phytohormones as potential positional cues varies depending on the species. Understanding precisely how these distinct polarity components interact to divide the small field of organ initials into adaxial and abaxial domains remains a major challenge facing the study of leaf polarity.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4655
References
- 1.McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 2.Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- 3.Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- 5.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 7.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 10.Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 11.Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nogueira FTS, Madi S, Chitwood DH, Juarez MT, Timmermans MC. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 2007;21:750–755. doi: 10.1101/gad.1528607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timmermans MC, Schultes NP, Jankovsky JP, Nelson T. Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development. 1998;125:2813–2823. doi: 10.1242/dev.125.15.2813. [DOI] [PubMed] [Google Scholar]
- 14.Juarez MT, Twigg RW, Timmermans MC. Specification of adaxial cell fate during maize leaf development. Development. 2004;131:4533–4544. doi: 10.1242/dev.01328. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H. The putative RNA dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell. 2005;17:2157–2171. doi: 10.1105/tpc.105.033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 17.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 18.Bhalerao RP, Bennett MJ. The case of morphogens in plants. Nat Cell Biol. 2003;5:939–943. doi: 10.1038/ncb1103-939. [DOI] [PubMed] [Google Scholar]
- 19.Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 20.Chitwood DH, Guo M, Nogueira FTS, Timmermans MC. Establishing leaf polarity: The role of small RNAs and positional signals in the shoot apex. Development. 2007;134:813–823. doi: 10.1242/dev.000497. [DOI] [PubMed] [Google Scholar]