Abstract
To defend themselves, plants activate inducible defense mechanisms that are effective against the invader that is encountered. There is partial overlap in the defense signaling pathways that are induced by insect herbivores and microbial pathogens that may result in cross-resistance. We have previously shown that infestation by tissue-chewing Pieris rapae larvae induces resistance in Arabidopsis thaliana against subsequent attack by the microbial pathogens Pseudomonas syringae pv. tomato (Pst), Xanthomonas campestris pv. armoraciae (Xca) and turnip crinkle virus (TCV). Phloem-feeding aphids, such as the generalist Myzus persicae, have a stealthy feeding strategy that is very different from chewing by lepidopteran larvae. Yet, M. persicae feeding results in a large transcriptomic change. Here, we report on the effectiveness of the defense response that is triggered by M. persicae infestation, as well as the sensitivity of M. persicae to microbially-induced resistance. M. persicae reproduction was not affected by prior conspecific feeding, nor was aphid-induced resistance effective against subsequent attack by Pst, Xca or TCV. Moreover, induced systemic resistance (ISR) triggered by beneficial Pseudomonas fluorescens rhizobacteria was not effective against M. persicae. However, systemic acquired resistance (SAR) induced by prior infection with avirulent Pst was associated with reduced aphid reproduction. These data provide insight into the effectiveness of pathogen and insect resistance and highlight the complexity of the defense responses that are triggered during multitrophic plant-attacker interactions.
Key Words: Arabidopsis, induced resistance, defense signaling, Myzus persicae
Plants are abundantly present on earth and are at the basis of almost all food webs. Plants face a multitude of attackers such as herbivorous insects and pathogenic microbes. While there are ca. 300,000 plant species, there are expected to be ca. Three million species of herbivorous insects.1 The diversity of pathogenic microbes is less well characterized but their threat to plants is equally renowned.2 To effectively combat invasion by pathogens and insects plants have evolved sophisticated strategies to “perceive” biotic interactions and to translate this “perception” into an appropriate defensive or conducive response.3–6 Recent genomics research revealed that the plant's capacity to respond to the enormous diversity of parasites and beneficials is highly flexible.7–12 Signaling networks that are recruited by the plant in response to pathogens and insects overlap, indicating that the regulation of the plant's adaptive response is finely-balanced between protection against microbial and insect aggressors.
To understand how plants integrate pathogen- and insect-induced signals into specific defense responses, we previously monitored the dynamic production of the defense signals salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), and performed large-scale gene expression studies in Arabidopsis thaliana upon attack by a set of microbial pathogens and herbivorous insects with different modes of attack.7 The results obtained in this study indicated that plants induce a highly attacker-specific gene expression pattern that is shaped, among other signals, by SA, JA and ET. Next, we investigated whether insect-induced resistance provides cross-resistance against pathogens in Arabidopsis. We demonstrated that resistance induced by larvae of the cabbage white butterfly Pieris rapae is not only effective against P. rapae itself, but also against several microbial pathogens.13 For instance, P. rapae feeding locally reduced symptoms caused by the bacterial pathogens Pseudomonas syringae pv. tomato (Pst) and Xanthomonas campestris pv. armoraciae (Xca). Moreover, P. rapae feeding induced local and systemic resistance against turnip crinkle virus (TCV).
In contrast to tissue maceration by larvae of P. rapae, aphids minimize damage to their hosts. They use their flexible stylets to probe intercellularly between the plant cells to reach the phloem and feed for a prolonged period of time from a single sieve element.14 These different feeding strategies result in very distinct gene expression patterns upon attack by Myzus persicae (green peach aphid) or P. rapae.7 Here, we investigated the spectrum of effectiveness of aphid-induced resistance against several microbial pathogens, including Pst, Xca and TCV. In addition, we studied microbially-induced resistance against M. persicae.
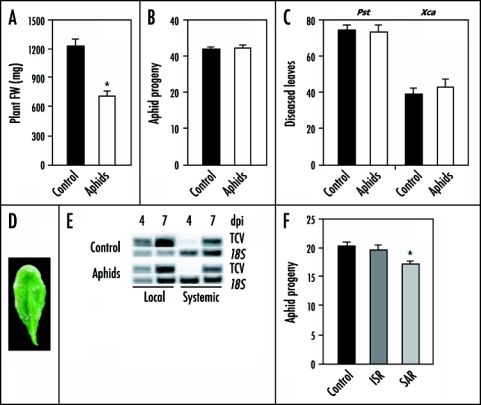
Previously, we demonstrated that three days of feeding by 40 aphids does not result in detectable increases in the levels of SA, JA or ET.7 Surprisingly, gene expression profiling of Arabidopsis after 48 and 72 hour of M. persicae infestation revealed that aphids induce a major reprogramming of the Arabidopsis transcriptome.7 The majority of the genes with changed expression are predicted to play a role in plant secondary metabolism. Figure 1A shows that aphid feeding resulted in a ∼40% reduction in plant fresh weight, presumably through removal of photoassimilates (carbon) and amino acids (nitrogen) from the phloem. Hence, the previously observed changes in the transcriptome may be caused by a shift in source-sink relations.
Figure 1.
Arabidopsis-Myzus persicae interactions. (A) Infestation of five-week-old Arabidopsis Col-0 plants with 20 adult aphids for three days significantly reduced the fresh weight (FW) of Arabidopsis rosettes (n = 20). (B) Progeny per adult aphid while feeding for seven days on un-induced Col-0 plants and on plants that had been pre-infested with aphids for three days (n = 15). (C) Percentage of leaves with disease symptoms caused by the bacterial pathogens P. syringae pv. tomato DC3000 (Pst) or X. campestris pv. armoraciae (Xca) in control and aphid-induced Col-0 plants (n = 15). (D) Aphid-vectored TCV symptoms on Arabidopsis Di-0 plants. (E) Local and systemic accumulation of TCV RNA at four and seven days post inoculation (dpi) in inoculated control and aphid-induced Col-0 plants. (F) The number of offspring produced per adult aphid while feeding for six days on uninduced Col-0 plants and on plants expressing P. fluorescens WCS417r-mediated ISR, or P. syringae pv. tomato DC3000(avrRpt2)-induced SAR (n = 20). Green peach aphids (Myzus persicae) were reared on radish (Raphanus sativus), in a greenhouse (25°C, RH 50–70%, L16:D8h). The 16 h light period prevented sexual reproduction, keeping the population clonal. A synchronous colony was started by transferring apterous adults to uninfested radish plants. The aphids were enclosed in clip-cages. After 24 h, the adults were removed and the newly born nymphs kept until they moulted. Using a fine paint brush, single, recently moulted (one to three days) adult apterae were transferred to single Col-0 plants. Each plant was confined in a Magenta GA-7 vessel. offspring per aphid was determined after six to seven days in a climate chamber (23°C, RH 70%, L16:D8h). For induction treatments, all aphids were removed after three days, after which the plants were challenged with fresh aphids or pathogens. Induction of ISR and SAR, and pathogen assays were performed as described (refs. 13,25). Student's t-test (A–C) or one-way ANoVA with a Bonferroni post-hoc test (F) (p < 0.05) was performed to test for significant differences. Statistically significant differences compared to the control are indicated with asterisks.
In order to investigate whether the response that is triggered in Arabidopsis upon feeding by M. persicae results in enhanced aphid resistance, we monitored the reproduction of this attacker. Arabidopsis accession Columbia-0 (Col-0) plants were infested with 20 aphids. After 72 hours, all aphids were removed and plants were infested with a single adult aphid. M. persicae reproduction was the same on pre-treated and control plants (Fig. 1B), indicating that Arabidopsis does not recognize the attacker or cannot mount an effective defense response against this attacker. The latter is more likely, as Arabidopsis plants showed large transcriptional changes upon M. persicae infestation. Also in potato, M. persicae feeding did not result in induced resistance to M. persicae.15 It is thought that aphids manipulate plant responses and thereby avoid or suppress induction of effective defense responses.16,17
Similar experiments were performed with the challenging pathogens Pst, Xca and TCV. Although the overlap in genes differentially expressed upon M. persicae and Pst attack was approximately 30%,7 prior feeding by aphids did not cause cross-resistance to either Pst or Xca (Fig. 1C). Green peach aphids are capable of transmitting over 100 virus species,18 including TCV. Hence, aphid feeding could serve as a cue for the plant to prepare for potential secondary viral infections. We hypothesized that prior feeding by M. persicae would lead to induced resistance against TCV. In Col-0 plants, TCV spreads systemically throughout the plant and does not produce visible disease symptoms. Accession Dijon-0 (Di-0) exhibits a hypersensitive response (HR) to TCV, resulting in confined nectrotic lesions.19–21 First, we tested if M. persicae is able to transmit TCV to non-infected tissue in Arabidopsis Di-0. Indeed, as a result of M. persicae feeding, TCV was vectored to uninfected leaves of Arabidopsis Di-0 plants where they cause HR-like symptoms (Fig. 1D). Next, M. persicae-induced resistance against TCV was tested in Col-0 by monitoring the amount of TCV RNA in local and systemic leaves at different time points after infection. Control and aphid-induced plants showed equal amounts of TCV RNA (Fig. 1E), indicating that the responses triggered in Arabidopsis by aphid feeding did not result in cross-resistance against TCV. Together, these results indicate that infestation of Arabidopsis by M. persicae does not result in enhanced resistance to the microbial pathogens Pst, Xca and TCV.
In reciprocal experiments, aphid reproduction was assessed after elicitation of microbially-induced resistance. Rhizobacteria-mediated induced systemic resistance (ISR),22 triggered by Pseudomonas fluorescens WCS417r,23 was not effective against M. persicae (Fig. 1F). However, systemic acquire resistance (SAR),24 triggered by infection with avirulent Pst,25 significantly reduced aphid reproduction. The number of aphid offspring on SAR-expressing plants was significantly reduced in comparison to control plants (Fig. 1F). Although it is unclear what signals contribute to this effective aphid control, it is likely that a rise in SA levels, which is typically associated with SAR, plays a dominant role in aphid resistance.26 Recently, it was shown in Arabidopsis that nymphs of the phloem-feeding insect Bemisia tabaci (Silverleaf Whitefly) sabotage effectual JA-dependent host defenses by activating the antagonistic SA signaling pathway.27 Since M. persicae has a similar feeding strategy, and the SA marker gene PR-1 is activated upon aphid feeding,7 the inhibitory effect of pathogen-induced SAR on aphid development may resemble this SA-mediated antagonistic effect. On the other hand, JA has also been implicated in resistance to M. persicae,28 and since JA is systemically produced upon elicitation of SAR by avirulent Pst,29,30 we can not exclude a role for JA in defense against M. persicae. These data show that a biotic interaction between a plant and one of its attackers may influence interactions with subsequent attackers.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4663
References
- 1.Schoonhoven LM, Van Loon JJA, Dicke M. Insect-Plant Biology. Oxford: Oxford University Press; 2005. [Google Scholar]
- 2.Agrios GN. Plant Pathology. San Diego: Elsevier Academic Press; 2005. [Google Scholar]
- 3.Dicke M, Hilker M. Induced plant defences: From molecular biology to evolutionary ecology. Basic Appl Ecol. 2003;4:3–14. [Google Scholar]
- 4.Slusarenko AJ, Fraser RSS, Van Loon LC. Mechanisms of Resistance to Plant Diseases. Dordrecht: Kluwer Academic Publishers; 2000. [Google Scholar]
- 5.Van Loon LC. Systemic induced resistance. In: Slusarenko AJ, Fraser RSS, Van Loon LC, editors. Mechanisms of Resistance to Plant Diseases. Dordrecht: Kluwer Academic Publishers; 2000. pp. 521–574. [Google Scholar]
- 6.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 7.De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC, Dicke M, Pieterse CMJ. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant-Microbe Interact. 2005;18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- 8.Verhagen BWM, Glazebrook J, Zhu T, Chang HS, Van Loon LC, Pieterse CMJ. The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant-Microbe Interact. 2004;17:895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- 9.Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou GZ, Katagiri F. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell. 2003;15:317–330. doi: 10.1105/tpc.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kempema LA, Cui X, Holzer FM, Walling LL. Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs: Similarities and distinctions in responses to aphids. Plant Physiol. 2007;143:849–865. doi: 10.1104/pp.106.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reymond P, Bodenhausen N, Van Poecke RMP, Krishnamurthy V, Dicke M, Farmer EE. A conserved transcriptional pattern in response to a specialist and a generalist herbivore. Plant Cell. 2004;16:3132–3147. doi: 10.1105/tpc.104.026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez L, Weidmann S, Arnould C, Bernard AR, Gianinazzi S, Gianinazzi-Pearson V. Pseudomonas fluorescens and Glomus mosseae trigger DMI3-dependent activation of genes related to a signal transduction pathway in roots of Medicago truncatula. Plant Physiol. 2005;139:1065–1077. doi: 10.1104/pp.105.067603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Vos M, Van Zaanen W, Koornneef A, Korzelius JP, Dicke M, Van Loon LC, Pieterse CMJ. Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiol. 2006;142:352–363. doi: 10.1104/pp.106.083907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tjallingii WF, Hogen Esch T. Fine structure of aphid stylet routes in plant tissues in correlation with EPG-signals. Physiol Entomol. 1993;18:317–328. [Google Scholar]
- 15.Alvarez AA. Resistance mechanisms of Solanum species to Myzus persicae. The Netherlands: Wageningen University; 2007. Ph.D. Thesis. [Google Scholar]
- 16.De Vos M, Kim JH, Jander G. Biochemistry and molecular biology of Arabidopsis-aphid interactions. Bioessays. 2007;29:871–883. doi: 10.1002/bies.20624. [DOI] [PubMed] [Google Scholar]
- 17.Will T, Tjallingii WF, Thonnessen A, Van Bel AJE. Molecular sabotage of plant defense by aphid saliva. Proc Natl Acad Sci USA. 2007;104:10536–10541. doi: 10.1073/pnas.0703535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blackman RL, Eastop VF. Aphids on the World's Crops: An identification and information guide. Chichester, UK: Wiley; 2000. [Google Scholar]
- 19.Simon AE, Li XH, Lew JE, Stange R, Zhang C, Polacco M, Carpenter CD. Susceptibility and resistance of Arabidopsis thaliana to turnip crinkle virus. Mol Plant-Microbe Interact. 1992;5:496–503. [Google Scholar]
- 20.Dempsey DA, Pathirana MS, Wobbe KK, Klessig DF. Identification of an Arabidopsis locus required for resistance to turnip crinkle virus. Plant J. 1997;11:301–311. doi: 10.1046/j.1365-313x.1997.11020301.x. [DOI] [PubMed] [Google Scholar]
- 21.Ton J, Van Pelt JA, Van Loon LC, Pieterse CMJ. Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol Plant-Microbe Interact. 2002;15:27–34. doi: 10.1094/MPMI.2002.15.1.27. [DOI] [PubMed] [Google Scholar]
- 22.Van Loon LC, Bakker PAHM, Pieterse CMJ. Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol. 1998;36:453–483. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- 23.Pieterse CMJ, Van Wees SCM, Hoffland E, Van Pelt JA, Van Loon LC. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell. 1996;8:1225–1237. doi: 10.1105/tpc.8.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 25.Pieterse CMJ, Van Wees SCM, Van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, Van Loon LC. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran PJ, Thompson GA. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001;125:1074–1085. doi: 10.1104/pp.125.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zarate SI, Kempema LA, Walling LL. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 2007;143:866–875. doi: 10.1104/pp.106.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis C, Karafyllidis L, Turner JG. Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol Plant-Microbe Interact. 2002;15:1025–1030. doi: 10.1094/MPMI.2002.15.10.1025. [DOI] [PubMed] [Google Scholar]
- 29.Pieterse CMJ, Van Pelt JA, Ton J, Parchmann S, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol Mol Plant Pathol. 2000;57:123–134. [Google Scholar]
- 30.Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci USA. 2007;104:1075–1080. doi: 10.1073/pnas.0605423104. [DOI] [PMC free article] [PubMed] [Google Scholar]