Abstract
Nep1-like proteins (NLPs) are a novel family of microbial elicitors of plant necrosis that induce a hypersensitive-like response in dicot plants. The spatial structure and role of these proteins are yet unknown. In a paper published in BMC Plant Biology (2008; 8:50) we have proposed that the core region of Nep1-like proteins (NLPs) belong to the Cupin superfamily. Based on what is known about the Cupin superfamily, in this addendum to the paper we discuss how NLPs could form oligomers.
Key words: quaternary structure, necrosis and ethylene inducing proteins, NLPs, MpNEP1, MpNEP2, NPP1, Moniliophthora perniciosa, Phytophthora parasitica
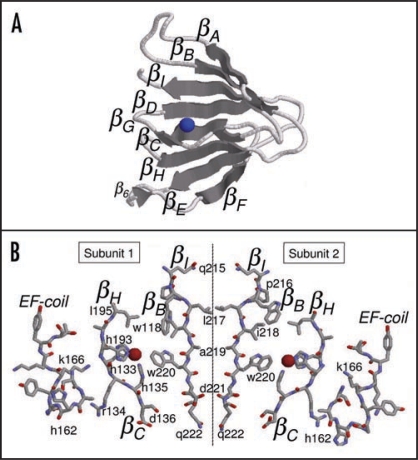
Cupins may be organized as monomers, dimers, hexamers and octamers of β-barrel domains.1 To the best of our knowledge trimers have not been detected yet. The interaction of two monomers building up a dimeric structure is basically performed by three types of interactions: hydrophobic interactions between β-strands in different subunits, salt bridges and hydrogen bonds between β-strands. In cupin dimers, the hydrophobic interactions occur between two βI strands in different subunits (Fig. 1A and B). This strand represents the central axis of rotation of the dimer as one residue in βI interacts with the corresponding residue in the other subunit (Fig. 1B). Therefore, all residues in βI must be hydrophobic, as one residue interacts with the other subunit and the next one in the sequence interacts with the interior of the protein. Charged residues in βI would disrupt such interactions. Most cupin dimers have strong hydrophobic residues such as tryptophan (W), phenylalanine (F) and methionine (M) pointing towards the own subunit (↓), while small hydrophobic residues such as leucine (L), isoleucine (I), and valine (V) point to the other subunit (↑). A particular case is leucine that interacts with other subunits, for instance, βI = liaW (positions 217–220 in Fig. 1B) and βI = LVsw of type I and II NLP consensuses, respectively. Therefore, the pattern of hydropathicity suggests that the side chain orientation is βI = l217 ↑ i218 ↓ a219 ↑ W220 ↓ d221 ↑. However we observe that just after βI there is a charged residue (aspartate D221) which would point outwards disrupting the dimer or at least making it less stable. It is interesting to observe that the requirement for a negatively charged residue at this last position is very high: 96% of all type I NLPs contains an aspartate (D) or glutamate (E) indicating an important role for it, maybe in avoiding dimerization of the NLPs. A second interesting hypothesis is as follows: several cupins are oxygenases, decarboxylases, etc. and use a negatively charged residue, such as aspartate or glutamate as proton donor.1 Now, if the alternate pattern of side chains of the residues is βI = l217 ↓ i218 ↑ a219 ↓ W220 ↑ d221 ↓, instead of the previous one, then the aspartate or glutamate residue would point to the hydrophobic pocket and would be positioned to interact with the metal ion, as in cupins with enzymatic activity. However, there are no experimental evidences that the NLPs have enzymatic activity.
Figure 1.
(A) Three-dimensional structure prediction for type I NLP consensus, (B) Interface between two βI strands in type I NLP consensus. From the left to the right: EF-coil with the conserved residue H162, βC and βH strands (superposed) with the conserved histidines H133 and H135 in βC, H193 and leucine L195 in βH, W220 in βI and W118 in βB. The strands in the right subunit follow the same pattern but rotated.
The second type of interaction is salt bridges between charged residues in different subunits. Analyzing all interacting side chains in the 1VJ2 protein (dimer), we verify that the charged side chains of N35 and E57 (numbers in original structure) are only 2.72 Å apart. In the NLPs, this corresponds to N10836% (Q10860%) at the border of βB and E13898%. The negatively charged residue D125 helps to correct the orientation of the subunits in relation to each other avoiding any disorientation. The high conservation level of these residues suggests that NLPs are dimeric structures. However, as we will see next, only hydrophobic and charged interactions are not enough to build a dimer.
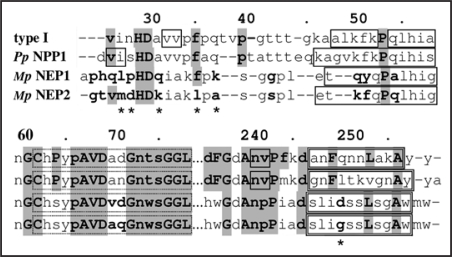
Garcia et al. (2007)2 have used small angle X-ray scattering (SAXS) to show that, in solution, at low concentrations (<2 mg/ml) the two copies of the NLPs of Moniliophthora perniciosa, MpNEP1 and MpNEP2, exist as dimers and monomers, respectively. The same technique showed that at higher concentrations, >5 mg/ml, both proteins exist as dimers, as is the case for PpNPP1.2 They also reported, based on electrophoresis analysis, that PpNPP1 and MpNEP1 exist as oligomers and MpNEP2 as monomers.2 However, experiments with the PpNPP1 in size exclusion chromatography using myoglobin as size standard suggest that PpNPP1 is a monomer.3 Figure 2 compares MpNEP1, MpNEP2 and PpNPP1, where the most relevant differences in sequence are marked with asterisks (*) and are possibly related to the differences in oligomeric properties between MpNEP1 and PpNPP1 with MpNEP2. These positions are methionine M27 and leucine L35, which occur only in MpNEP2, glycine G250, which occurs only in MpNEP2 and NEP1 (Fusarium oxysporum) and lysine K31, which occurs only MpNEP2, BAB04114 (Bacillus halodurans) and AAU23136 (Bacillus licheniformis). The other residues are aspartate D28, which occurs 9 times and alanine A37 which occurs 7 times of all investigated NLPs. Thus, the sequence mdHDkiakl at the start of the NLPs seems to explain the monomeric state of MpNEP2, although at higher concentrations they form dimers. Besides the weak hydrophobic interactions, dimeric cupins and bicupins (two β barrels in the same sequence building up a dimeric-like 4d-structure) are stable structures (see Fig. 1 in ref. 4). By aggregating the first β-strand in the start domain of one β-barrel to the ABIDG β-sheet of the other β-barrel, composing a big ABIDGY β-sheet (Y is the first β-strand). For instance, using the bicupin 1L3J (oxalate decarboxylase) as template, the low confidence level β-strand at position 26–33 (v in H29D30 avv) in type I NLPs corresponds to the first β-strand. Since this proceeds from both barrels they can build a stable structure (see Fig. 1 in ref. 4). The quaternary structure is related to the presence of interaction residues in the BID β-sheet of the cupin structure. These are present in the NLPs and would enable them to form dimers.
Figure 2.
Alignment of type I NLP consensus, PpNPP1, MpNEP1 and MpNEP2. Solid line boxes are β-strands, double line boxes are α-helices. The sequence positions marked with asterisks (*) are possibly related to the differences in oligomeric properties between MpNEP1 and PpNPP1 with MpNEP2.
Acknowledgments
Work partially supported by CNPq grants 506414/2004-3, 151171/2007-6 and FAPESP grant 2007/02827-9.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/6643
References
- 1.Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Garcia O, Macedo J, Tibórcio R, Zaparoli G, Rincones J, Bittencourt L, Ceita G, Micheli F, Gesteira A, Mariano A, Schiavinato M, Medrano F, Meinhardt L, Pereira G, Cascardo J. Characterization of necrosis and ethylene-inducing proteins (NEP) in the basidiomycete Moniliophthora perniciosa, the causal agent of witches' broom in Theobroma cacao. Mycol Res. 2007;111:443–455. doi: 10.1016/j.mycres.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, Engelhardt S, Felix G, Kemmerling B, Krzymowska M, Nürnberger T. NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 2002;32:375–390. doi: 10.1046/j.1365-313x.2002.01454.x. [DOI] [PubMed] [Google Scholar]
- 4.Cechin AL, Sinigaglia M, Lemke N, Echeverrigaray S, Cabrera OG, Pereira GAG, Mombach JCM. Cupin: A candidate molecular structure for the Nep1-like protein family. BMC Plant Biol. 2008;8:50. doi: 10.1186/1471-2229-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]