Abstract
PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2 are two negative regulators of shade avoidance syndrome (SAS) responses in Arabidopsis. PAR1 and PAR2 belong to the bHLH family of transcription factors and act as direct transcriptional repressors of auxin- and brassinosteroid-responsive genes. These observations led us to propose that PAR1 and PAR2 might integrate shade and hormone signals. After plant proximity perception by the phytochrome photoreceptors, the expression of PAR1, PAR2 and dozens of additional PAR genes is affected, initiating a complex web of transcriptional events instrumental for the establishment of the SAS responses. Studying the organization of this complex transcriptional network, that is, the interactions amongst the different PAR factors involved and how they are connected with the endogenous hormone-regulated transcriptional networks, seems therefore fundamental to understand how SAS is modulated.
Key words: arabidopsis, auxin, bHLH, PAR genes, phytochrome, regulatory modules, SAUR genes, shade avoidance
Presence of nearby plants results in a reduction in the red to far-red ratio (R:FR) of the incoming light, a signal perceived by the phytochrome photoreceptors that triggers rapid and reversible changes in dozens of PHYTOCHROME RAPIDLY REGULATED (PAR) genes.1–6 Several of these genes are likely instrumental for establishing the SAS responses. Among them we have focused on PAR1 and its paralogous PAR2, that encode proteins related in sequence to the bHLH family of transcription factors with no previous assigned function.3 To investigate the role of PAR1 and PAR2 in planta we followed a reverse genetics approach. The phenotypic analyses of plants with increased or reduced PAR1 and/or PAR2 levels suggested a negative role for these genes in the SAS regulation, indicating that altered levels of PAR1 and/or PAR2 significantly affect plant responsiveness to simulated shade.
The similarity of PAR1 and PAR2 to members of the bHLH family of transcription factors led us to think that they could regulate development by modulating gene transcription. In agreement with this idea, (i) fusions of PAR1 and PAR2 to GFP reporter proteins showed that both are nuclear proteins, and (ii) constitutive overexpression of a chimera of PAR1 and the glucocorticoid receptor (GR) in plants induced a severe dwarf dark-green phenotype only in the presence of the synthetic glucocorticoid dexamethasone.3 A more in depth analysis of their primary sequence revealed that PAR1 and PAR2 lack consensus sequences for DNA binding,7,8 suggesting that they would function as transcription cofactors.9 As such, they would modulate the DNA binding activity of other factors by protein-protein interactions probably through the HLH domain, as proposed for other non DNA binding bHLH proteins in plants or animals.10–12 As a consequence, PAR1 and PAR2 might interact with many different bHLH factors acting as integrators of various signals. In that context, identification of PAR1 partners will allow us to unveil new SAS gene regulators that might also be early targets of phytochrome action (Fig. 1).3 Indeed, other identified SAS regulators, such as HFR1 and PIL12,13 also encode members of the bHLH family. Additionally, several PAR genes encode for bHLH proteins.1,2,14 The functional analyses of different PAR genes, such as HFR1, PAR1, PAR2 and PIL1, suggest the prevalence of negative factors among the PAR members with a demonstrated role in controlling SAS responses.2,3,6,13 Nevertheless, a few positive regulators of SAS, such as ATHB2, PIF4 and PIF5, have also been described.15,16 Understanding the molecular interactions between these regulators, i.e., if they are organized in regulatory modules17 controlling distinct circuits of the shade-regulated transcriptional network involved in implementing different SAS responses, appears as fundamental to understand how the SAS is regulated.
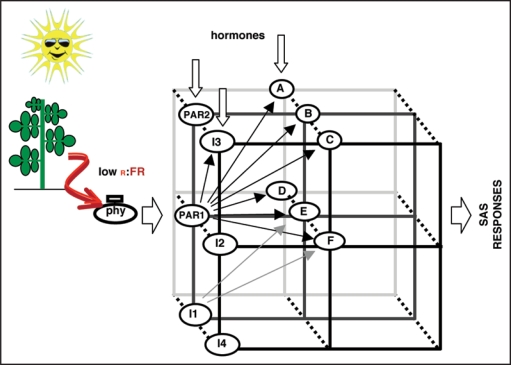
Figure 1.
Simplified scheme showing PAR1 and PAR2 in the transcriptional network used by phytochromes to induce SAS responses upon plant proximity perception. PAR1 and PAR2 have a role in this network, together with other PAR factors, like ATHB2, HFR1 and PIL1. Some of these factors might be interacting with PAR1 and PAR2. As a result, different PAR factors might be organized in regulatory modules, integrated within this web. We propose that PAR1 and PAR2 would connect light signals with hormonal cues, since SAUR15 and SAUR68, identified as direct targets of PAR1 action, are also hormone regulated. The identification of additional partners of PAR1 and/or PAR2 and their direct targets will help to fill gaps in this complex web of interactions. I1 to I4: partners of PAR1 and/or PAR2. (A–F) direct targets of PAR1 and/or PAR2; (E and F) SAUR15 and SAUR68 respectively.
Analysis of global transcript profiles of wild type and PAR1 overexpressing plants led to the identification of SMALL AUXIN UPREGULATED15 (SAUR15) and SAUR68 as direct targets of PAR1 action. These two genes belong to a gene family already described as upregulated by auxins18 and/or brassinosteroids (BR).19 Despite the broad evidence in support of the interplay between light and hormonal signaling,20 data on the precise molecular links between SAS and hormonal transcriptional networks are scarce. Our results represent a direct link between plant proximity perception by the phytochromes and auxin and BR signaling, and postulate these two atypical bHLH proteins as novel regulators directly integrating shade and hormone transcription networks (Fig. 1).
As expected for direct targets of PAR1, SAUR15 and SAUR68 are also regulated by simulated shade (Fig. 1). However, the shade regulation of SAUR15 and SAUR68 expression can not be explained solely by the regulatory action of PAR1. This is not surprising, since in the context of a transcriptional network, direct targets of PAR1 are very likely shared by other PAR1 partners. The identification of additional direct targets of PAR1 can therefore be helpful to establish a map of interactions between PAR1, PAR2 and other factors, providing proof of the concept that the shade-modulated transcriptional web controlling SAS is organized in regulatory modules.
Overall, our results have demonstrated that these atypical bHLH proteins, belonging to the subgroup VIII-A of bHLH proteins,7 negatively control plant growth and metabolic SAS responses.3 They act in the nucleus impairing the auxin-regulated expression of SAUR15 and SAUR68 genes and functioning as transcriptional cofactors that would rapidly connect shade perception by phytochromes and hormone responsiveness and/or sensibility, integrating the corresponding transcriptional networks. In the future, we will focus on PAR1 interactors and their direct targets in order to unravel new components of the complex transcriptional network initiated by the phytochromes upon onset of SAS. Only those direct targets that are also shade-regulated will be considered as true targets of PAR1 and PAR1 interactors.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5599
References
- 1.Devlin PF, Yanovsky MJ, Kay SA. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 2003;133:1617–1629. doi: 10.1104/pp.103.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426:680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- 3.Roig Villanova I, Bou Torrent J, Galstyan A, Carretero Paulet L, Portoles S, Rodriguez Concepcion M, Martinez Garcia JF. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 2007;26:4756–4767. doi: 10.1038/sj.emboj.7601890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carabelli M, Morelli G, Whitelam G, Ruberti I. Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA. 1996;93:3530–3535. doi: 10.1073/pnas.93.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I. The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J. 1993;4:469–479. doi: 10.1046/j.1365-313x.1993.04030469.x. [DOI] [PubMed] [Google Scholar]
- 6.Roig Villanova I, Bou J, Sorin C, Devlin PF, Martinez Garcia JF. Identification of Primary Target Genes of Phytochrome Signaling. Early Transcriptional Control during Shade Avoidance Responses in Arabidopsis. Plant Physiol. 2006;141:85–96. doi: 10.1104/pp.105.076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 8.Toledo Orti G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 10.Hyun Y, Lee I. KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol. 2006;61:283–296. doi: 10.1007/s11103-006-0010-2. [DOI] [PubMed] [Google Scholar]
- 11.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 12.Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113:3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 13.Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005;19:2811–2815. doi: 10.1101/gad.364005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carabelli M, Possenti M, Sessa G, Ciolfi A, Sassi M, Morelli G, Ruberti I. Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 2007;21:1863–1868. doi: 10.1101/gad.432607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development. 1999;126:4235–4245. doi: 10.1242/dev.126.19.4235. [DOI] [PubMed] [Google Scholar]
- 16.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 17.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 18.McClure BA, Guilfoyle T. Characterization of a class of small auxin-inducible soybean polyadenilated RNAs. Plant Mol Biol. 1987;9:611–623. doi: 10.1007/BF00020537. [DOI] [PubMed] [Google Scholar]
- 19.Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halliday KJ, Fankhauser C. Phytochrome-hormonal signaling networks. New Phytol. 2003;157:449–463. doi: 10.1046/j.1469-8137.2003.00689.x. [DOI] [PubMed] [Google Scholar]