Abstract
In Arabidopsis, the CLE genes encode a family of at least 32 peptide ligands. Our gain-of-function studies demonstrated that all of the 18 genes we examined caused pleiotropic and often opposing phenotypes, including various combinations of increased root and rosette growth, root stunting, dwarfing, shoot apical meristem (SAM) arrest, asymmetric leaf development, and “shrublike” phenotypes. Many CLE genes caused similar phenotypes that correlated with common amino acid substitutions among subsets of the genes, suggesting key amino acids necessary for certain phenotypes. The pleiotropic phenotypes we observed were the results of integrated hypermorphic and global neomorphic responses to abundant ectopic ligands through multiple signaling pathways. The phenotypes are also suggestive of wide ranging, often antagonistic roles played by these genes in plant development. The interpretations of our findings and some apparently contradictory recent results are discussed in this context.
Key words: arabidopsis, clavata, CLE, hypermorphy, neomorphy, peptide ligand, plant development
The CLE gene family consists of 32 known members in Arabidopsis.1–4 However, the developmental processes regulated by these genes are known for but a few, with demonstrated roles in SAM maintenance,1,5 root nutation6 and tracheary element differentiation.4 Although the probable functional segments of the CLE proteins are post-translationally cleaved and modified dodecapeptides,4,7 27 distinct dodecapeptide sequences are found in this family, suggesting additional functions in plant developmental processes. Effects of CLE genes on root development1,3,8–10 and varied locations of expression6,9,11 also support this notion.
To further address the roles played by this large family of ligands, we conducted a gain-of-function study via gene overexpression of 18 CLE family members.3 The CLE overexpression phenotypes were pleiotropic and suggested roles in root and shoot growth, phyllotaxis, leaf size and shape, apical dominance and developmental timing. Similarities in phenotypes, which enabled their categorization into four broad classes, suggested possible functional redundancies among the CLE genes. Key functional amino acid residues were suggested by strong correlations of certain phenotypes with specific, tandem substitutions in CLE domain amino acid sequences. For example, the CLE41, 42 and 44 genes identified in our study were unique in having the combination of the three amino acids his, ile and ser at positions 3, 12 and 13 in the CLE domain (numbering as in ref. 3), which perfectly correlated with the distinctive “shrub-like” phenotype observed for the plants that overexpressed these genes (ref. 3, unpublished results).
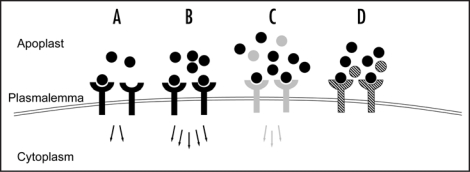
The pleiotropic phenotypes we observed are perhaps best understood in the context of hypermorphy and neomorphy, key considerations in the interpretation of phenotypes in any gain-offunction experiment (reviewed in ref. 12). Hypermorphy can be described as an amplification of the normal phenotypic effect that a protein has on an organism (Fig. 1). In contrast, neomorphy is a novel phenotype that would not normally be caused by a protein. In peptide ligand overexpression, neomorphy could result from abnormal signaling due to interactions with receptors in cells where the ligand would not normally be found. Alternatively, overexpression could raise a ligand's concentration well above a non-cognate receptor's dissociation constant for the ligand, thereby resulting in a signal that would not normally occur due to low binding affinity (Fig. 1). Finally, antimorphy (dominant negative neomorphy) could potentially result from a ligand binding a receptor in a way that blocks signal transduction. If antimorphy occurs in CLE overexpression, it would most likely be through competitive inhibition, given the sequence similarities amongst the CLE peptides (Fig. 1).
Figure 1.
Ligand overexpression-mediated hypermorphy, neomorphy and antimorphy. (A) Normal ligand-receptor signal transduction in which the ligand (black circles) interacts with its cognate receptor (black “goblets”) at a physiologically normal effective concentration, with its concomitant signal transduction (black arrows); (B) Hypermorphic response, in which receptor binding sites are more heavily occupied due to the higher effective ligand concentration, with a concomitant increased signal; (C) Neomorphic response, in which the overexpressed ligand (black circles) interacts with a non-cognate receptor (gray “goblets”) to transduce a stronger than normal signal (gray arrows). The cognate ligand (gray circles) can still interact with the receptor and thereby contributes to the total signal strength (D) Antimorphic response, in which the ectopic ligand interacts with a non-cognate receptor (cross-hatched “goblets”), but in a manner that does not transduce a signal, thereby inhibiting signal transduction from the cognate ligand (cross-hatched circles). A competitive inhibition model is shown here, but other mechanisms of inhibition are possible.
Most, if not all, of the phenotypes from our CLE gene overexpression work are pleiotropic and thus likely neomorphic in some respect. For example, the SAM arrest phenotypes we observed for CLEs 2, 3, 4, 5, 6, 7 (Category Aii) and CLEs 9, 10, 11 and 13 (Category Ai)3 are probably neomorphic, since there is no evidence that any ligand other than CLV3 is required for signal transduction through the CLV1/2 complex.
An interesting aspect of our findings was the apparently opposite phenotypes that were observed amongst the CLE gene overexpressers. While nearly all the CLE overexpressers were dwarf in stature, CLE26 overexpressers were significantly larger than wild-type by 21 days after germination and CLE18 did not affect plant rosette area. Likewise, CLE9, 10, 11, 13, 19, 21 and CLV3 caused root stunting, while CLE2, 4, 5, 6, 7, 18, 25 and 26 stimulated root elongation. These results suggest that the CLE genes play antagonistic roles to each other in plant growth and development. Surprisingly, the exogenous peptide application study of Kinoshita et al.,13 in which Arabidopsis plants were grown on agar media or liquid media containing 31 of the 32 Arabidopsis CLE peptides (CLE43” was omitted), yielded many SAM and root phenotypes that appeared to contradict our observations resulting from CLE gene overexpression.3 For example, in roots, Kinoshita et al., either observed no difference (CLE2 and 4–7) or even inhibition of elongation (CLE18, 25 and 26)13 with CLEs that we found to be stimulatory to root elongation. Conversely, we observed no change in root growth with CLE42 and 44 overexpression, while Kinoshita et al.,13 observed slight root growth stimulation with in the presence of these peptides.
The apparent contradictions between these two studies are likely explained by differences in experimental approach. In our study, the strong “constitutive” CaMV 35S promoter was used, so not only were the ligands abundantly expressed, most were likely expressed in tissues and cells where they would not normally be found. Thus, the ligands would have been available to interact with receptors throughout the plant at higher than normal concentrations. The phenotypes we observed were thus likely the result of the integration of hypermorphic and multiple neomorphic signals through multiple receptors. Such integrated responses would presumably also be occurring with exogenous CLE peptides. However, these responses would presumably only be from the receptors present at (or very near to) the epidermal cells, which would therefore only be a subset of the receptors that would have been affected in our overexpression studies. Further work is needed to clarify the reasons for these differences. Even so, the results of Kinoshita et al.,13 confirm our observation that CLE peptides appear to play opposing roles to one another in plant growth and development.3
Irrespective of whether the CLE overexpression and exogenous application phenotypes are hypermorphic or neomorphic, they provide valuable clues to the developmental processes that CLE's help regulate. The characterisation of the gain-of-function phenotypes of the remaining CLE genes in Arabidopsis will thus provide additional useful data. Ultrastructural phenotypic analysis will further aid in understanding the developmental roles played by these peptides. RNAi approaches may also yield useful data, but this could prove difficult, given the low transcript abundances from these genes1,3,6,11 and possible functional redundancies. Given the many apparently opposing phenotypes observed in overexpression and exogenous application studies, it appears that the CLEs may often function antagonistically to one another in plant growth and developmental processes such as root elongation. It will be of great interest to determine if these antagonisms constitute components of dynamic feedback loops that regulate developmental processes. The challenge will be in separating the hypermorphic from the neomorphic, connecting each ligand to its “true” phenotype, receptor and thereby, developmental process.
Acknowledgements
I thank Anne-Marie Smit, Philip O'Donnell and Elspeth MacRae for helpful discussions and comments on the manuscript.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5602
Note added in proof
Ogawa et al., (Science 2008; 319:294) demonstrated that the binding affinities of CLV3 and CLE peptides to CLV1 are consistent with our observations of the strength of CLV3's hypermorphic or the CLE peptides' neomorphic SAM phenotypes conferred by ectopic expression of the CLE ligands.3
References
- 1.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 2.Cock JM, McCormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001;126:939–942. doi: 10.1104/pp.126.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strabala TJ, O'Donnell PJ, Smit AM, Ampomah Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen N, Quinn B, Foote HCC, Hudson KR. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 2006;140:1331–1344. doi: 10.1104/pp.105.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 5.Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JV. CLV3 is localized to the extracellular space, where it activates the arabidopsis CLAVATA stem cell signaling pathway. Plant Cell. 2002;14:969–977. doi: 10.1105/tpc.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobe M, Müller R, Grünewald M, Brand U, Simon R. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Gen Evol. 2003;213:371–381. doi: 10.1007/s00427-003-0329-5. [DOI] [PubMed] [Google Scholar]
- 7.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 8.Casamitjana Martinez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol. 2003;13:1435–1441. doi: 10.1016/s0960-9822(03)00533-5. [DOI] [PubMed] [Google Scholar]
- 9.Fiers M, Hause G, Boutilier K, Casamitjana Martinez E, Weijers D, Offringa R, van der Geest L, van Lookeren Campagne M, Liu CM. Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene. 2004;327:37–49. doi: 10.1016/j.gene.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 10.Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell. 2005;17:2542–2553. doi: 10.1105/tpc.105.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma VK, Ramirez J, Fletcher JC. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol. 2003;51:415–425. doi: 10.1023/a:1022038932376. [DOI] [PubMed] [Google Scholar]
- 12.Zhang JZ. Overexpression analysis of plant transcription factors. Curr Opin Plant Biol. 2003;6:430–440. doi: 10.1016/s1369-5266(03)00081-5. [DOI] [PubMed] [Google Scholar]
- 13.Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. Gain-of-Function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:1821–1825. doi: 10.1093/pcp/pcm154. [DOI] [PubMed] [Google Scholar]