Abstract
Arabidopsis has Aux/IAA proteins that lack domain II which is a binding site for the TIR1 auxin receptor. These proteins have been shown to be more stable than the canonical Aux/IAA proteins. We investigated the phenotypes of overexpression lines of domain II-less Aux/IAA proteins, IAA20 and IAA30, by the use of 35S promoter. The transgenic lines showed many aberrant phenotypes in auxin physiology. The most conspicuous phenotype was collapse of the root apical meristem as occurred in plt1 plt2 double mutants. Because IAA20 and IAA30 were early auxin-inducible and were expressed in the root apical meristem, they may have a physiological role in maintaining the stem cell niche of root, by keeping the activity of MP/ARF5 and NPH4/ARF7 at an acceptable level. On the other hand, domain-II less Aux/IAA proteins are not present in balck cottonwood tree or grapevine, suggesting wide diversification of Aux/IAA proteins in higher plants.
Key words: auxin signal transduction, Aux/IAA protein, protein degradation, root apical meristem
Aux/IAA genes play a pivotal role in auxin signal transduction since their products have been shown to be a direct target of the auxin F-box receptors including TIR1, which are components of SCF ubiquitin ligase E3 complexes.1,2 TIR1 binds the degron sequence of Aux/IAA proteins3 that is conserved in most of them and is called domain II since their finding. Importance of domain II is apparent because amino acid substitutions in its core sequence cause dominant mutations, which are supposed to occur due to accumulation of the mutated protein. However, 5 of the 29 Arabidopsis Aux/IAA proteins such as IAA20 and IAA30, lack domain II. Another Aux/IAA protein, IAA31, has a G-to-D substitution in the degron sequence, which is similar to a G-to-E substitution in a dominant mutation of an Aux/IAA gene, iaa3/shy2.4 Consistent with these characteristics of domain II, IAA20 and IAA31 have been shown to be long-lived compared to the canonical IAA17 protein.5
Because the physiological role of these noncanonical Aux/IAA proteins is unknown, we made transgenic Arabidopsis lines overexpressing IAA20, IAA30 or IAA31 cDNA by means of 35S promoter, and investigated their phenotype. The three gene products are most related each other in amino acid sequence and form a single clade in the Arabidopsis Aux/IAA proteins.6 Each overexpression line (OX) showed qualitatively similar defects in auxin physiology, with IAA20 OX being the most severely affected. IAA20 OX showed low fecundity, low germination rate, short and agravitropic hypocotyls and shoots, agravitropic roots and defects in formation of vasculature. Most interestingly, growth of the primary roots was arrested soon after germination due to collapse of the root apical meristem (RAM). Lateral roots were formed and grew with fewer occasions of collapse than the primary roots. We also found that IAA20 and IAA30 were early auxin inducible, but IAA31 was not induced by auxin. The collapse of RAM observed in the OXs is similar to that in plt1 plt2 double mutants.7 PLTs encode AP2-class transcription factors, and are late auxin-inducible through the function of ARF5/MP and ARF7/NPH4. They act to establish the stem cell niche of root, and are essential for the root stem cell activity.7,8 Therefore, accumulated IAA20 proteins may inhibit these ARFs, which would result in repression of PLT expression and ultimately bring about severe reduction of meristematic activity in roots of OX.
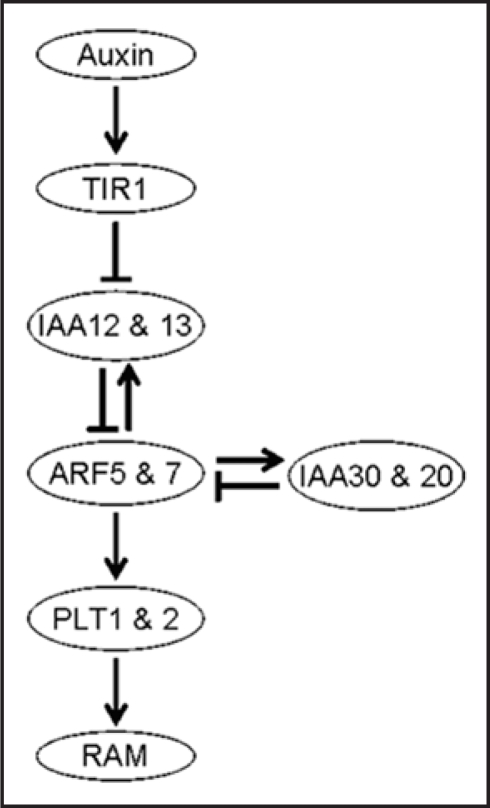
Recently IAA30 has been shown to be specifically expressed in the quiescent center cells (QC) of RAM. IAA20 is also expressed in the columella root cap and QC, but IAA31 was expressed at a low level without specificity in RAM.9 Using IAA20 or IAA30 promoter-GUS reporter genes, we also confirmed the expression of both genes in QC and the columella root cap (unpublished data). On the other hand, MP activity, through which PLTs are transcribed, is regulated through twin canonical Aux/IAA proteins, IAA12/BDL and IAA13 in RAM formation during embryogenesis.10,11 Therefore, we propose an auxin signaling pathway that acts to maintain the stem cell niche of root as shown in Figure 1. Because IAA20 and IAA30 do not contain domain II, they should not act directly downstream of TIR1. Furthermore, we postulate that they are auxin-inducible through the function of MP and NPH4. Then, what is the role of IAA20 and IAA30, which can not interact with the TIR1 auxin receptor? Ectopic expression of PLT in the embryo causes homeotic transformation of the apical region to root and hypocotyl identity,7 suggesting that strict control of their expression is needed for correct formation and maintenance of RAM. Because IAA20 and IAA30 are not degraded through the function of the TIR1 auxin receptor, their levels tend to remain the same, and may only increase in response to auxin. These characteristics of IAA20 and IAA30 suggest that they act as damper in the auxin signaling pathway that lessen the fluctuations of the MP and NPH4 levels and ensures that expressions of the late auxin-inducible PLT genes remain at acceptable levels. Recently IAA30 has been also reported to express through a transcription factor, LEC2 and has been suggested to be involved in somatic embryogenesis.12,13 Though the DNA binding domain of LEC2 (B3 domain) is related to that of ARFs,14 it binds to the RY motif, which is different from the auxin response element.12
Figure 1.
Model for domain II-less Aux/IAA proteins (IAA30 and IAA20) in maintenance of the root apical meristem (RAM).
We investigated the presence of the noncanonical Aux/IAA proteins in four other plant species for which complete genome sequences were available (Table 1). Each has Aux/IAA proteins with the dominant mutant-type domain II. Domain II-less Aux/IAA proteins also often lack domain I. Black cottonwood tree and grapevine do not have Aux/IAA proteins that lack only domain II. Thus, domain II-less Aux/IAA proteins like IAA20 or IAA30 of Arabidopsis are found only in Arabidopsis and rice. OsIAA8 (Os02g49160) of rice appears to be an orthologue of IAA20 or IAA30 of Arabidopsis.15 However, it is not auxin-inducible.16 OsIAA4 (Os01g13030) has the dominant mutant-type domain II like Arabidopsis IAA31. However, unlike IAA31, it is auxin-inducible.16 Moss Physcomitrella patens does not have any noncanonical Aux/IAA proteins. These results suggest that Aux/IAA proteins diversified extensively in both structure and function in higher plants. Thus, no consistent roles could be assigned to the noncanonical Aux/IAA proteins at present.
Table 1.
Noncanonical Aux/IAA proteins in a few plant species
| Species | Total number of Aux/IAA proteins | Domain II-less proteins | Dominant mutation-type proteins* | References |
| Arabidopsis thaliana | 29 | 5 (1)** | 1 (0)** | 5 |
| Oryza sativa | 31 | 5 (3) | 1 (0) | 15***, 16 |
| Populus trichocarpa | 35 | 3 (3) | 2 (0) | 15 |
| Vitis vinifera | 31 | 6 (6) | 3 (1) | This study |
| Physcomitrella patens | 2 | 0 | 0 | This study |
Aux/IAA proteins with the domain II sequences which are comparable to those of dominantly-mutated Aux/IAA proteins of Arabidopsis.
Atypical Aux/IAA proteins often also lack domain I. Numbers in parenthesis show proteins without domain I and domain II.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5994
References
- 1.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 2.Kepinski S, Leyser O. The Arabidopsis TIR1 protein is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 3.Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanismof auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 4.Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- 5.Dreher KA, Brownn J, Saw RE, Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell. 2006;18:699–714. doi: 10.1105/tpc.105.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004;135:1738–1752. doi: 10.1104/pp.104.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 9.Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005;24:1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. PNAS. 2006;103:3468–3473. doi: 10.1073/pnas.0511331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc Natl Acad Sci USA. 2008;105:3151–3156. doi: 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 15.Kalluri UC, DiFazio SP, Brunner AM, Tuskan GA. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007;7:59. doi: 10.1186/1471-2229-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa) Funct Integ Genomics. 2006;6:47–59. doi: 10.1007/s10142-005-0005-0. [DOI] [PubMed] [Google Scholar]