Abstract
Pre-harvest sprouting (PHS) leads to loss of grain weight and a reduction in the end use quality of kernels in cereals, especially in wheat, and PHS in rice also becomes a more and more serious problem recent years. Many factors are involved in the controlling this complex trait. Only recently, we have reported the large scale screening and charactersation of the rice phs mutants, providing insight into the molecular mechanism of pre-harvest sprouting in rice. It has been shown that mutations of genes in synthesis of the carotenoid precursors of ABA resulted in the pre-harvest sprouting, which is consequence of ABA deficiency, and photobleaching is likewise due to the absence of photoprotective carotenoids. The further study of all different rice phs mutants will help us to elucidate the complex phenomena and finally capture the target for improving PHS in rice or other cereals.
Key words: pre-harvest sprouting, ABA, carotenoids, rice
Introduction
The phenomenon of germination of physiologically mature cereal grains in the ear or panicle, usually under wet conditions shortly before harvest, is termed as pre-harvest sprouting (PHS) or vivipary. PHS occurs in many cereal crops such as wheat, barley, maize, and rice in most region of the world. PHS not only causes reduction of grain yield, but also affects the quality of grains, resulting into significant economic losses.
During seed formation, embryo development can proceed through a maturation phase that allows the entry into a quiescent state, characterized by acquisition of desiccation tolerance, growth arrest and the entry into a dormancy period of variable length that is broken upon germination.1 It is known that the plant hormone abscisic acid (ABA) is strongly involved in this process, but the mechanism is still not fully understood. It was found that ABA levels are low during embryogenesis, increase during the maturation phase, and then decrease when seed desiccation. So far a number of mutants with reduced capacity to synthesize ABA have been described, such as flc, not and sit in tomato; aba1 in Nicotiana plumbaginifolia; aba1, aba2 and aba3 in Arabidopsis; and viviparous mutants Vp5, Vp7, Vp10/Vp13, Vp14, Vp15 in maize.2–4 Most viviparous mutants in maize were blocked in biosynthesis of the carotenoid precursors for de novo ABA synthesis; these mutants produce albino or pale green, non-viable seedlings.5 However, some mutants in Arabidopsis involved in the carotenoid biosynthetic pathway did not lead to precocious germination.6–8 In rice, relatively few phs mutants have been reported so far, which only two genes related to vivipary were cloned, without further detail study.9 Recently, we have carried out an intensive screening of the rice mutant population and successfully isolated 27 phs mutants under the high humidity paddy fields.10 This review focuses on more recent studies of phs or viviparous mutants, and discusses the complex regulation of ABA synthesis and its physiological role in seed dormancy and germination.
Viviparous or Pre-Harvest Sprouting Mutants in Crops
Since pre-harvest sprouting in wheat is very intricate, most viviparous genes were identified through isolation of the mutants from other cereal crops, especially in maize. At least seven viviparous genes in maize including Vp1, Vp5, Vp7, Vp8, Vp10/Vp13, Vp14, Vp15 were cloned.2–4 The Vp5 and Vp7 genes encode enzymes in the carotenoid biosynthetic pathway, and the mutants showed an albino phenotype with a reduced ABA level.11,12 The Vp14 gene, on the other hand, is blocked in the first committed step in ABA biosynthesis, cleavage of epoxy-carotenoids to xanthoxin.13,14 The Vp10/Vp13 and Vp15 encode the enzymes in the molybdenum cofactor biosynthesis, the final step for ABA biosynthesis, oxidation of ABA-aldehyde to ABA by an aldehyde oxidase which requires molybdenum cofactor (MoCo). Vp1 encodes B3 domain transcription factors, which controls multiple developmental responses associated with the maturation phase of seed formation.15,16 Vp8, encoding a putative altered meristem program1-like peptidase, regulates ABA accumulation and coordinates embryo and endosperm development.17 In addition, some other mutant loci in maize were identified in the carotenoid biosynthetic pathway, such as Y1, Vp2, W3, Vp9 and Y9.11,18–20
Carotenoids and phs Mutant in Rice
Recently we have identified a series of rice phs mutants simply categorized into three groups based on phenotypes besides vivipary, and the genes in four different loci were cloned in category I, that are all located in the carotenoid biosynthetic pathway.10 Unlike the viviparous mutants of maize, which accumulate carotenoids mainly in the endosperm, the rice counterparts accumulated carotenoids in seedling or embryo. It is thought that PSY, catalyzing the first committed step in carotenogenesis, is rate-limiting at least in many non-green tissues.21 There are three PSYs in rice, which OsPSY1 and OsPSY2 involved in carotenoid biosynthesis in photosynthetically active tissues, while OsPSY3 is devoted to abiotic stress-induced ABA formation. Interestingly, the transcripts for all three PSYs were not detected in rice endosperm.22 Therefore, the carotenoids do not accumulate in the endosperm of rice phs mutants. To this end, it is not surprising that we didn't identify any psy mutants in our large scale screening for phs mutants simply due to gene redundancy.
Carotenoids are integral and essential components of the photosynthetic membranes in all plants. In the chloroplast, they function in the protection against photo-oxidative damage and participate in the light harvesting process.23 The rice phs mutants in the carotenoids biosynthetic pathway showed chloroplast damages, and phs1, phs2 and phs4 mutants are albino and lethal. Interestingly, the phs3 mutant can survive and showed ‘variegated’ leaf at the tillering stage and completely leaf photobleaching during grain filling. The β-OsLCY RNAi plants also showed photobleaching leaf, and the levels of some PS II core proteins decreased in the plants.10 These results are consistent with the conclusion that carotenoids protect against oxidative damage.
ABA Biosynthesis and Pre-Harvest Sprouting
ABA is involved in several specific processes during seed development, such as the deposition of storage reserves, induction of primary dormancy. Evidence for the role of ABA in such processes has come from ABA-deficiency or -response mutants in Arabidopsis or maize.24 Mutations in ABA biosynthesis fail to induce seed dormancy and exhibit a vegetative wilty phenotype, such as Arabidopsis aba1 and tobacco aba2 are known to be impaired in ZEP (zeaxanthin epoxidase), the first enzyme identified as an ABA biosynthetic enzyme;25 The Arabidopsis ABA-deficient mutant aba4 was recently identified in a screening for paclobutrazol-resistant germination and showed impairment in NSY (neoxanthin synthase).26 The maize viviparous14 (vp14) and tomato notabilis mutants are shown to be defective in NCED, 9-cis-epoxycarotenoid dioxygenase catalyzing the oxidative cleavage of xanthophylls, 9-cis-violaxanthin and/or 9′-cis neoxanthin to produce xanthoxin.14,27 Arabidopsis aba2 and aba3, maize vp10 and vp15, and tomato flacca and sitiens are typical mutants impaired in the later steps of ABA biosynthesis in the cytosol.28–31 By comparison, ABA-deficiency or -response mutants in rice were scarcely identified. At present we have cloned other three genes from phs mutants besides PHS1–PHS4 genes, and they are all involved in specific ABA biosynthetic pathway. Further detailed characterization with these mutants is underway.
ABA Response and Pre-Harvest Sprouting
Thus far, our knowledge on the signaling elements that mediate the regulation of seed dormancy and germination by ABA is primarily derived from genetic analysis. The maize Vp1 locus was the first cloned gene in ABA response, and has been studied in detail.16 Vp1 is a multidomain transcription factor that functions as both an activator and a repressor depending on the promoter context.15 Interestingly, the missplicing of wheat Vp1 genes and rice Vp1 counterpart contributes to susceptibility to PHS in modern hexaploid wheat varieties and the sprouting susceptible rice varieties, respectively.32,33 ABI3 is orthologous gene of VP1 from Arabidopsis, vp1 and abi3 seeds share similar phenotypes including insensitivity to ABA, desiccation intolerance and premature activation of the shoot apical meristem.34 Mutations in the ABI4 and ABI5 loci have similar ABI3 qualitative effects on seed development and ABA sensitivity, but null mutations in ABI3 are more severe than those in ABI4 or ABI5.35
In addition, several other loci in Arabidopsis have been identified that specifically affect seed maturation and germination but do not appear to be directly related to hormone synthesis or signalling. The leafy cotyledon (LEC) class genes, including LEC1, LEC2 and FUS3,36 play key regulatory roles in Arabidopsis affecting important traits of the maturation phase during seed development and the establishment and maintenance of dormancy.1 However, very few information about the homologous genes in cereals was reported, which are worth for further studying.
Conclusions and Perspectives
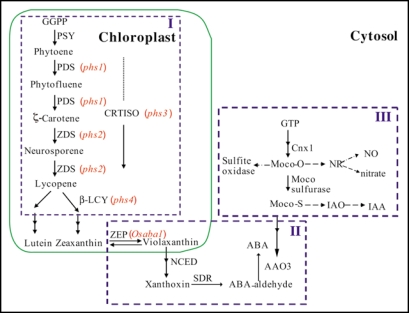
Recent advances in ABA biosynthesis research have yielded substantial information on the pathways, genes and enzymes involved in the process.37 We have learned that ABA is the major hormone involved in induction and maintenance of dormancy by pre-harvest sprouting or viviparous mutants in rice. Based on these results, the phs mutants in cereals are involved within three parts in the ABA biosynthesis pathway (Fig. 1). Part I represents carotenoids biosynthesis, maize vp5, vp7 and rice phs1–psh4 are localized in this part. Part II represents specific ABA biosynthesis, this part mutants include maize vp14, rice Osaba1. Part III is involved in molybdenum cofactor biosynthesis, maize vp10/vp13 and vp15 are in that part. We are going to identify the phs mutants involved in all three parts. Identification of downstream targets of ABA and the genes that regulate ABA biosynthesis will help us to gain deep understanding on the dormancy mechanisms of cereals, and how ABA represses germination and prevent pre-harvest sprouting in crops.
Figure 1.
Pre-harvest sprouting mutants in rice localized in carotenoid and abscisic acid biosynthetic pathway. (I) Carotenoid precursor synthesis in the early steps of ABA biosynthesis. (II) Specific ABA biosynthetic pathway. (III) Molybdenum cofactor biosynthesis, molybdenum cofactor is a factor for active AAO3. GGPP, Geranylgeranyl pyrophosphate; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, χ-carotene desaturase; β-LCY, lycopene β-cyclase; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; SDR, short-chain dehydrogenase/reductase; AAO3, Abscisic aldehyde oxidase 3; IAO, Indole-3-acetaldehyde oxidase; NR, Nitrate reductase; Cnx1, cofactor for nitrate reductase and xanthine dehydrogenase 1.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/6606
References
- 1.Vicente-Carbajosa J, Carbonero P. Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol. 2005;49:645–651. doi: 10.1387/ijdb.052046jc. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz SH, Qin X, Zeevaart JA. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes and enzymes. Plant Physiol. 2003;131:1591–1601. doi: 10.1104/pp.102.017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarty DR. Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:71–93. [Google Scholar]
- 4.Bentsink L, Koornneef M. Seed Dormancy and Germination. Rockville, MC: American Society of Plant Biologists; 2003. [Google Scholar]
- 5.Seo M, Koshiba T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002;7:41–48. doi: 10.1016/s1360-1385(01)02187-2. [DOI] [PubMed] [Google Scholar]
- 6.Qin G, et al. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid and gibberellin biosynthesis. Cell Res. 2007;17:471–482. doi: 10.1038/cr.2007.40. [DOI] [PubMed] [Google Scholar]
- 7.Giuliano G, Giliberto L, Rosati C. Carotenoid isomerase: a tale of light and isomers. Trends Plant Sci. 2002;7:427–429. doi: 10.1016/s1360-1385(02)02329-4. [DOI] [PubMed] [Google Scholar]
- 8.Dong H, et al. The Arabidopsis Spontaneous Cell Death1 gene, encoding a zeta-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Res. 2007;17:458–470. doi: 10.1038/cr.2007.37. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal GK, et al. Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol. 2001;125:1248–1257. doi: 10.1104/pp.125.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang J, et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 2008;54:177–189. doi: 10.1111/j.1365-313X.2008.03411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hable WE, Oishi KK, Schumaker KS. Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol Gen Genet. 1998;257:167–176. doi: 10.1007/s004380050636. [DOI] [PubMed] [Google Scholar]
- 12.Singh M, et al. Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell. 2003;15:874–884. doi: 10.1105/tpc.010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 14.Tan BC, Schwartz SH, Zeevaart JA, McCarty DR. Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoecker U, Vasil IK, McCarty DR. Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev. 1995;9:2459–2469. doi: 10.1101/gad.9.20.2459. [DOI] [PubMed] [Google Scholar]
- 16.McCarty DR, et al. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki M, et al. The maize Viviparous8 locus, encoding a putative ALTERED MERISTEM PROGRAM1-like peptidase, regulates abscisic acid accumulation and coordinates embryo and endosperm development. Plant Physiol. 2008;146:1193–1206. doi: 10.1104/pp.107.114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buckner B, Miguel PS, Janick-Buckner D, Bennetzen JL. The y1 gene of maize codes for phytoene synthase. Genetics. 1996;143:479–488. doi: 10.1093/genetics/143.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews PD, Luo R, Wurtzel ET. Maize phytoene desaturase and zeta-carotene desaturase catalyse a poly-Z desaturation pathway: implications for genetic engineering of carotenoid content among cereal crops. J Exp Bot. 2003;54:2215–2230. doi: 10.1093/jxb/erg235. [DOI] [PubMed] [Google Scholar]
- 20.Li F, Murillo C, Wurtzel ET. Maize Y9 encodes a product essential for 15-cis-zeta-carotene isomerization. Plant Physiol. 2007;144:1181–1189. doi: 10.1104/pp.107.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diretto G, et al. Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS ONE. 2007;2:350. doi: 10.1371/journal.pone.0000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsch R, Wust F, Bar C, Al-Babili S, Beyer P. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 2008;147:367–380. doi: 10.1104/pp.108.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demmig-Adams B, Gilmore AM, Adams WW., 3rd Carotenoids 3: in vivo function of carotenoids in higher plants. FASEB J. 1996;10:403–412. doi: 10.1096/fasebj.10.4.8647339. [DOI] [PubMed] [Google Scholar]
- 24.Holdsworth M, Kurup S, Mkibbin R. Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci. 1999;4:275–280. [Google Scholar]
- 25.Marin E, et al. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996;15:2331–2342. [PMC free article] [PubMed] [Google Scholar]
- 26.North HM, et al. The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 2007;50:810–824. doi: 10.1111/j.1365-313X.2007.03094.x. [DOI] [PubMed] [Google Scholar]
- 27.Burbidge A, et al. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 1999;17:427–431. doi: 10.1046/j.1365-313x.1999.00386.x. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz SH, Leon-Kloosterziel KM, Koornneef M, Zeevaart JA. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997;114:161–166. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porch TG, Tseung CW, Schmelz EA, Mark Settles A. The maize Viviparous10/Viviparous13 locus encodes the Cnx1 gene required for molybdenum cofactor biosynthesis. Plant J. 2006;45:250–263. doi: 10.1111/j.1365-313X.2005.02621.x. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki M, et al. The maize viviparous15 locus encodes the molybdopterin synthase small subunit. Plant J. 2006;45:264–274. doi: 10.1111/j.1365-313X.2005.02620.x. [DOI] [PubMed] [Google Scholar]
- 31.Sagi M, Scazzocchio C, Fluhr R. The absence of molybdenum cofactor sulfuration is the primary cause of the flacca phenotype in tomato plants. Plant J. 2002;31:305–317. doi: 10.1046/j.1365-313x.2002.01363.x. [DOI] [PubMed] [Google Scholar]
- 32.McKibbin RS, et al. Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc Natl Acad Sci USA. 2002;99:10203–10208. doi: 10.1073/pnas.152318599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan J, et al. Short, direct repeats (SDRs)-mediated post-transcriptional processing of a transcription factor gene OsVP1 in rice (Oryza sativa) J Exp Bot. 2007;58:3811–3817. doi: 10.1093/jxb/erm231. [DOI] [PubMed] [Google Scholar]
- 34.Giraudat J, et al. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meinke DW, Franzmann LH, Nickle TC, Yeung EC. Leafy cotyledon mutants of Arabidopsis. Plant Cell. 1994;6:1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]