Abstract
Polyamines have long been implicated in plant growth and development, as well as adaptation to abiotic and biotic stress. As a general rule of thumb the higher the polyamine titers the better. However, their molecular roles in plant stress responses still remain obscure. It has been postulated that they could act through their catabolism, which generates molecules which may act as secondary messengers signalling networks of numerous developmental and stress adaptation processes. Recently it was shown that plant and mammalian polyamine catabolism share critical features, giving new insight in plant polyamine catabolism. In this review, the advances in genes and proteins of polyamine catabolism in plants is presented and compared to other models.
Key words: polyamines, polyamine catabolism, polyamine oxidase, abiotic stress, ROS signaling
Introduction
Sensu stricto polyamines (PAs) are defined as the molecules that contain more than one amino groups. Sensu lato the term refers mostly to the widely found biogenic polycationic molecules, namely Putrescine (Put), Spermidine (Spd) and Spermine (Spm), with aliphatic structure found in all cells across all kingdoms. Cadaverine is also present in legumes, while thermospermine, a Spm isomer, is widespread in bacteria and higher plants.1 Since Spm is a general constituent of eucaryotic cells, but not of procaryotes, the identification of specific Spm functions in nuclei-containing cells was, and still is, a serious concern.
Plant PAs have been suggested to play important roles in divergent processes, such as gene expression, protein and DNA synthesis, cellular homeostasis, cell division and differentiation, growth and developmental processes such as embryogenesis, organogenesis, senescence, and also responses to abiotic and biotic stresses.2–9 Plant PAs have also been proposed to be responsible for characteristics of agro-economical importance, including phytonutrient content, fruit quality and plant life-span.10,11 Recently, it was shown that PAs are implicated in cell migration in mammals.12
Polyamine Homeostasis
Since PAs are implicated in such a divergent array of processes, their intracellular titers must be strictly regulated. Thus, apart from the rate of biosynthesis, the intracellular concentrations of free PAs are regulated by conjugation either to small molecules, especially hydroxycinnamic acids (soluble conjugated PAs),13–15 or with high molecular mass substances, like hemicelluloses and lignin and, to a lesser extent, proteins (so-called insoluble conjugated PAs).16 In addition to conjugation, the levels of free PAs can be downregulated by oxidative deamination. Cytoplasmic levels of PAs can be regulated also by subcellular compartmentalization to vacuoles, mitochondria and chloroplasts as well as by extrusion.14,17 In mammalian cells cultured under normal growth conditions, up to 90% of Put and 25% of Spd synthesized by the cells was secreted into the culture medium,18 while a similar trend has been observed in plant cells.19
Polyamine biosynthetic pathway is rather short, and the first PA to be synthesized is Put, via the Arg decarboxylase (ADC, EC 4.1.1.9), or Orn decarboxylase (ODC, EC 4.1.1.17) pathway, using Arg and Orn as substrates, respectively. Put is subsequently converted to Spd via Spd synthase (SPDS, EC 2.5.1.16) and Spd to Spm via Spm synthase (SPMS, EC 2.5.1.22), by sequential addition of an aminopropyl group. The aminopropyl group donor is decarboxylated S-adenosyl-L-methionine (dcSAM) produced by S-adenosyl-L-methionine decarboxylase (SAMDC, EC 4.1.4.50). SAM is a key intermediate for ethylene. Thus, antagonism between synthesis of higher PAs and ethylene may exist, since they share the same intermediate. These enzymes are under feedback control by their end products.20
Interestingly, the sequenced genome of Arabidopsis thaliana does not contain a gene encoding for ODC.21 So far, absence of this enzyme has only been reported in the protozoan eukaryote Trypanosoma cruzi. PAs synthesis in animals also starts from Orn, and can be formed directly from Pro by Orn aminotransferase.22 The ADC pathway via agmatine, as described for bacteria and plants, has not been demonstrated in animal cells. The existence of two pathways in plants for Put synthesis adds up significant complexity in PA homeostasis. Recently, a reconstruction of the evolutionary pathways of genes involved in PA biosynthesis23 provided plausible interpretations for the structural features that distinguish the different enzyme activities.
On the other hand, the main PA catabolic process is exerted through diamine oxidases (DAO, EC 1.4.3.6) and polyamine oxidases (PAO, EC 1.5.3.3), the former showing strong preference for diamines (Put and Cad), while the latter oxidize only higher PAs, such as Spd and Spm.24 DAOs are copper-containing proteins, with the tendency to form homodimers. They catalyze the oxidation of Put to 4-aminobutanal with concomitant production of NH3 and H2O2, and the resulting aldehyde is further converted to γ-aminobutyric acid (GABA) via Δ1-pyrroline. In addition, each subunit contains a 2,4,5-thrihydroxyphenylalanine quinine (TPQ) cofactor. In plants, DAOs occur at high levels in dicots, particularly pea, chickpea, lentil and soybean seedlings, loosely associated to cell wall.
Polyamine Oxidases
The first PA-catabolizing enzymes, which were characterized were DAOs25,26 and ruminant serum amine oxidase (SAO). The PAOs, able to catalyze the oxidative deamination of PAs with more than two amino-groups, bear FAD with non-covalent bonds that use N1-acetylderivatives as substrates in mammals, and non-acetylated PAs in plants.27 They are highly expressed in monocots, and can be classified as (i) those involved in the terminal catabolism of PAs, (ii) those involved in back-conversion, and (iii) those that contain a PAO domain but do not deaminate PAs.
From the first group, the best characterized enzyme so far is the maize PAO (ZmPAO). It is a 53 kD monomeric glycoprotein associated with cell walls and catalyzes the terminal catabolism of PAs, able to deaminate Spd and Spm, using FAD as cofactor and O2 as electron donor; the catabolic products are 4-aminobutanal or (3-aminopropyl)-4-aminobutanal, respectively, along with 1,3-diaminopropane and H2O2.
The second group, which resembles the mammalian PAOs, includes peroxisomal PAOs involved in a sequential back-conversion pathway of diacetylSpm (N1–N12) to acetylSpd and acetylSpd to acetylPut.28,29 Thus, in mammals an intriguing point is the absence of terminal catabolic PAOs. In addition, Spm oxidase (SMO, EC 1.5.3.3), a FAD-dependent amine oxidase was initially identified in animal cells; it catalyzes the back-conversion of Spm to Spd with concomitant production of 3-aminopropanal and H2O2. Recently an isoform was identified in the nucleus.24 Arabidopsis genome contains five putative PAO genes. Tavladoraki et al.30 identified PAO1 as an enzyme catalyzing the same reaction as SMO. Later, Moschou et al.31 identified PAO3 as the enzyme that completes the back-conversion pathway, converting Spm to Spd and Spd to Put. More importantly, in mammals PAOs that fall in this category are constitutively expressed, while their supply with their substrates depends on Spd/Spm N1-acetyltransferase (SSAT; EC 2.3.1.57) activity.24 On the other hand, SMO seems to be inducible, by certain polyamine analogues which adds considerably to the importance of this new pathway member because its regulation may contribute to the facilitation of tumor cell apoptosis.24 Furthermore, Helicobacter pylori induced SMO has been associated with increased ROS production and DNA damage, linking SMO to chronic inflammation and epithelial carcinogenesis, and also to prostate cancer.24
Moreover, a third class of enzymes includes lysine-decarboxylases-demethylases that possess also an amine oxidase domain similar to that of PAOs. These enzymes specifically localize to the nucleus and they are involved in the histone remodelling, the process of post-translational modifications of histone N-terminal tails.32 Within eukaryotic cells, DNA is packed into the higher order structure known as chromatin. The basic repeating unit of chromatin is the nucleosome, consisting of a histone octamer containing two copies each of histones H2A, H2B, H3 and H4, around which approximately 147 bp of DNA is wrapped. The histones of the histone octamer contain unstructured N-terminal “tails”, the residues of which are the site of numerous posttranslational modifications, including acetylation of lysine, phosphorylation of serine and threonine, and methylation of lysine and arginine. Additionally, lysine residues may be mono-, di- or trimethylated, while arginine residues may be mono- or dimethylated. The resulting complexity of modifications has been postulated to act as a “histone code”, by which these patterns of modifications are “read” by the cellular machinery to produce a specific gene regulatory outcome. The process impacts chromatin structure. This third class includes the LSD (lysine demethylase) proteins and requires FAD for catalytic activity.32
This category of enzymes is common between plants and animals, and the protein consists of two additional individual domains, except that of PAO, namely the SWIRM (Swi3p, Rsc8p and Moira), found in proteins involved in chromatin modifications or remodeling, and the spacer region.32 LSD1 functions as a transcriptional co-repressor through histone dimethylase activity specifically to histone H3 lysine 4, which is linked to active transcription. Interestingly, the PAO domain does not seem to exert any effect upon PAs. A. thaliana homologs of LSD1 promote floral transition through repressing the expression of floral repressor genes.33
In animals, PA catabolism requires the highly regulated inducible enzyme SSAT,34,35 which localizes to the cytoplasm and catalyzes the formation of N1-acetyl derivatives by the transfer of the acetyl group from acetylcoenzyme A to the N1 position of Spm/Spd. The acetyl derivatives are then converted to Spd or Put along with the production of 3-aceto-aminopropanal and H2O2 by the peroxisomal and constitutive PAO.27 It is worth noticing that in plants, PAO3 was also shown to be highly expressed (constitutive), whereas PAO1 was shown to be expressed at very low levels.30,31
Interestingly, mice chronically treated with the PAO inhibitor MDL72527 [N1,N4-bis(2,3-butadienyl)-1,4-butanediamine] died, due to Spm accumulation in red blood cells and blood plasma. This result indicates that blockage of PA catabolism, especially Spm catabolism, can be fatal in these animals,36 and emphasizes the importance of PA catabolism in mammals.
Polyamine Oxidases and Development
PAs are essential for growth and development, as inhibition of PA biosynthesis blocks cell growth.37–43 A reasoning of Spd requirement for eukaryotic viability has been recently established. Spd is a precursor of the unusual amino acid hypusine, which is involved in posttranslational modification of the ε-amino group of one specific lysine residue of the eukaryotic translational initiation factor 5A (eIF5A).44,45
In plants, PAOs, that are able to determine at least partly Spd titers, are abundant in lignifying tissues, and are spatially and temporally associated with cell wall strengthening reactions involving cell wall peroxidases.46 Also, PA catabolism has been associated with cell survival and cell growth, such as cell wall stiffening and lignification.46,47 PAO is involved in mesocotyl growth and in this context a progressive cellular re-distribution occurs toward primary and secondary cell walls during tissue maturation or light exposure (reviewed in ref. 47). This re-distribution probably affects tissue maturation by an increment in H2O2 production since the PAs titers are higher in cellular compartment than in the apoplast were maize PAO is localized. Furthermore, endogenous cues controlled the transcript levels of PAO in tobacco48 and in maize auxin suppressed whereas light upregulated expression of PAO.49
On the other hand, developmental programmed cell death (PCD) is preceded by an increase in PAO levels and a concomitant increase in H2O2.48 In tobacco, high PAs titers were associated with increased cell proliferation, whereas PAO was associated with leaves undergoing developmental PCD; also, PAO and PAs followed an inverse gradient; young tobacco leaves had increased PA content, while old leaves exhibited increased PAO activity.48 The considerable high ZmPAO in differentiating tracheary elements strengthens the view that PAOs are probably directly involved in PCD.47
These results suggest that PAOs are associated mostly with older rather than young and fast growing tissues, implying its involvement in maturation and secondary cell wall strengthening.
Polyamine Oxidases and Stress
PAs frequently accumulate in plants in response to abiotic and biotic stresses.5,50–52 There is extensive literature describing the correlation between endogenous PA titers and physiological perturbations and on the protective effect of PAs against environmental stresses (reviewed in refs. 7, 8 and 53). Exogenously supplied PAs protected plants from abiotic stress,54 whereas transgenic plants overexpressing PA biosynthetic genes exhibited stress tolerance.55–58 On the other hand, loss-of-function mutant of PA biosynthetic genes, or decrease of PA titers, resulted to decreased stress tolerance.59–63
PAs also upregulate stress-protective genes probably through their catabolism. For example, the tobacco ZFT1 is a Spm-responsive gene encoding for a zinc-finger type transcriptional repressor; tobacco plants overexpressing ZFT1 were more resistant to tobacco mosaic virus (TMV) compared to control plants.64 Furthermore, transgenic pine plants overexpressing the gene CaPF1, which encodes for the ERF (ethylene responsive factor)/AP2-type transcription factor, exhibited dramatically increased tolerance to drought, freezing and salt stress.65 In control pine plants, the levels of PAs decreased upon exposure to stresses, whereas in the transgenics plants the PA levels remained constant.
Recently, Moschou et al.,20 suggested that a crucial point in plant stress tolerance is the ratio of PA catabolism to PA anabolism. This could explain why overexpression of the gene CaPF1 resulted to increased tolerance by maintaining PA titers. Thus, enhanced stress tolerance in transgenic pines expressing CaPF1 was associated with high PA biosynthesis.66 In addition, apoplastic PA catabolism by PAO resulted to induction of stress-responsive genes, correlating PA catabolism to specific gene transcription.20
Moreover, Spm and Spd were potent inducers of nitric oxide (NO) in A. thaliana, but Put and its biosynthetic precursor Arg did not.67 NO inhibits oxidative phosphorylation in plant mitochondria68 and plays an important signalling role in plant-pathogen interactions.69 Therefore, further research on the role of PA-catabolism mediated NO production is justified.70
In mammals, the activation of PA catabolism increased cellular oxidative stress through the generation of H2O2, and induced the death of multiple types of cancer cells.30 Hydrogen peroxide, or/and the molecules produced through PAOs action, could exert signalling effects. In particular, 4-aminobutanal can be further metabolized to GABA, through the action of an aldehyde dehydrogenase (ADH, EC 1.2.1.3). GABA can also be produced by cytosolic glutamate dehydrogenase, and it is an important metabolite associated with various stress responses, including the regulation of cytosolic pH, carbon fluxes into the citric acid pathway, insect deterrence, protection against oxidative stress and signalling. DAP, on the other hand, is involved in tolerance since it is precursor of β-alanine and uncommon PAs.
A nodal point in stress responses is the activation of downstream responsive partners, such as Mitogen Activated Protein Kinases (MAPK) signalling pathways. H2O2 produced by PAO could trigger activation of MAPKs.71 Interestingly, PAO genes in Arabidopsis are upregulated by elicitors, such as flagellin, thus promoting probably activation of MAPKs involved in pathogen signal perception, while abiotic stresses also result to moderate increase.
Mechanical wounding stress significantly increased expression of PAO genes.31 Also, Tisi et al.72 provided evidence for the involvement of maize PAO to the wound healing response and to pathogen attack and more specifically to TMV. Yoda et al.73,74 reported that in Nicotiana tabacum plants resistant to TMV, PAO expression and PA titers increased in tissues exhibiting TMV-induced HR. Cell death caused by TMV infection or cryptogein, an oomycete-originated elicitor, was partially mediated by H2O2 generated through PA catabolism. These authors suggested that the PA substrate for H2O2 production is Spd, since during HR elicitation Spd but not Spm accumulated in the apoplast. That plants employ polyamine catabolism-derived H2O2 as a defensive tool upon exposure to biotic stresses has been reviewed by other groups.47,51,52
Moreover, tobacco PAO was an early-responsive gene when tobacco plants were challenged with the biotrophic Pseudomonas syringae pv tabaci (Moschou PN and Sarris et al., unpublished data). On the other hand, Takahashi et al.75 provided evidence for the correlation of Spm oxidation and induction of HR-associated and defense related genes. In addition, tolerance to TMV infection by treatment with Spm was modulated independently of SA.75 Finally, PAO RNAi lines showed significantly reduced PCD rate when treated with cryptogein.74 Whether PA accumulation under biotic stress is necessary to provide the catabolic pathway with substrate remains to be elucidated.
With respect to abiotic stress, Moschou et al.20 showed that upon salt stress Spd was secreted into the apoplast were it was catabolized by PAO producing H2O2. The titers of the apoplastic H2O2 was PAO-dependent. High PAO levels resulted to high H2O2 accumulation which induced PCD, whilst moderate levels of PAO and thus H2O2 resulted to the signalling and expression of defense genes, facilitating abiotic responses.20
Going Backwards with PAOs
Traditionally, plant PA catabolism was considered as a fundamentally divergent process when compared to the mammalian counterpart. Recently, Tavladoraki et al.,30 identified an A. thaliana PAO (AtPAO1) which possesses back-conversion activity, responsible for the conversion of Spm to Spd. Recently, Moschou et al.,31 identified the second PAO being able to back-convert, again in A. thaliana (AtPAO3), catalyzing the sequential conversion of Spm to Spd and Spd to Put. Interestingly, AtPAO3 was shown to be sorted into peroxisomes, the organelle in which mammalian PAOs are localized. More recently, Kamada-Nobusada et al.76 identified AtPAO2 and AtPAO4 to be sorted into peroxisomes. Moreover, AtPAO4 was shown to posses Spm and not Spd oxidase activity, similar to AtPAO1, thus back-converting Spm to Spd. Also, a PAO from barley (BPAO2) was the first identified plant PAO to have a terminal C-extension, thus a non-apoplastic localization, responsible for sorting to the vacuole.77
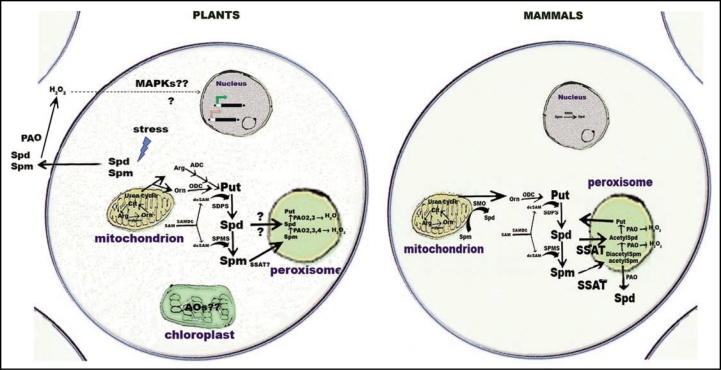
So far, the physiological roles of these PAOs remain far from understood, although AtPAO3 was shown to be induced by a wide-array of environmental stimuli involving both plant-pathogen and abiotic interactions. Moreover, the back-conversion pathway in plants most likely is independent of SSAT enzymes since all PAOs characterized in A. thaliana so far show strong preference for non-acetylated PAs, unlike mammalian ones that depend on SSAT pathway. What is worth noticing is the existence of biochemical data, which show the back-conversion of Spd to Put in other plant species, including Vitis vinifera plants (Toumi, Moschou PN and Roubelakis-Angelakis KA, unpublished data). Whether the pathway is only restricted to dicot plants remains to be identified. A comparative model for PA catabolism in plants and animals is summarized in Figure 1.
Figure 1.
Model for PA catabolism in plant and mammalian cells. In plants, peroxisomes and apoplast are the sites of PA oxidation by either back-convertion or terminal oxidation, respectively. Apoplastic PA oxidation has been correlated with the regulation of gene expression [either induction of defense genes (green) or genes related to PCD (red)] via redox signals during stress, while the role for peroxisomal PA oxidation remains obscure. Moreover, it seems that the plant peroxisomal PAO oxidation is SSAT-independent. The pathway that correlates the apoplastic redox state to the regulation of the gene expression is unknown. PAO2 and PAO4 were recently found to be localized to peroxisomes (Kamada-Nobusada et al., 2008) In mammals, PAOs are localized to peroxisomes, while SMOs are localized to the cytoplasm and to the nucleus. In addition, in mammals SMOs are found in the cytoplasm and in the nucleus.
Conclusion
In conclusion, recent data support the involvement of PAO genes in developmental and stress interactions. The picture has been complicated by the existence of additional PAO genes, with novel subcellular localizations and able to back-convert PAs instead of just being involved in their terminal catabolism. The recent availability of transgenic plants with up and downregulated the PAO genes and the respective mutants will greatly contribute to further reveal the physiological functions of these genes.
Abbreviations
- ROS
reactive oxygen species
- PAs
polyamines
- Put
putrescine
- Spd
spermidine
- Spm
spermine
- ADC
arginine decarboxylase
- ODC
ornithine decarboxylase
- SAMDC
S-adenosyl-L-methionine
- SPDS
spermidine synthase
- SPMS
spermine synthase
- DAO
diamine oxidase
- PAO
polyamine oxidase
- FAD
flavin adenine dinucleotide
- SSAT
Spd/Spm N1-acetyltransferase
- DAP
1,3-diaminopropane
- MAPK
mitogen activated protein kinases
- SMO
spermine oxidase
- LSD1
lysine demethylase 1
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7172
References
- 1.Mattoo AK, Handa AK. Higher polyamines restore and invigorate metabolic memory in ripening fruit. Plant Sci. 2008;174:386–393. [Google Scholar]
- 2.Kumar A, Altabella T, Taylor MA, Tiburcio AF. Recent advances in polyamine research. Trends Plant Sci. 1997:124–130. [Google Scholar]
- 3.Walden R, Cordeiro A, Tiburcio AF. Polyamines: small molecules triggering pathways in growth and development. Plant Physiol. 1997;113:1009–1013. doi: 10.1104/pp.113.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malmberg RL, Watson MB, Galloway GL, Yu WL. Molecular genetic analysis of plant polyamines. Crit Rev Plant Sci. 1998;17:199–224. [Google Scholar]
- 5.Bouchereau A, Aziz AF, Martin-Tanguy J. Polyamines and environmental challenges: recent development. Plant Sci. 1999;140:103–125. [Google Scholar]
- 6.Liu K, Fu H, Bei Q, Luan S. Inward potassium channel in guard cells as a target for polyamine regulation of stomatal movements. Plant Physiol. 2000;124:1315–1325. doi: 10.1104/pp.124.3.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alcázar R, Marco F, Cuevas JC, Patron M, Ferrando A, Carrasco P, Tiburcio AF, Altabella T. Involvement of polyamines in plant response to abiotic stress. Biotech Lett. 2006;28:1867–1876. doi: 10.1007/s10529-006-9179-3. [DOI] [PubMed] [Google Scholar]
- 8.Groppa MD, Benavides MP. Polyamines and abiotic stress: recent advances. Amino Acids. 2007;34:35–45. doi: 10.1007/s00726-007-0501-8. [DOI] [PubMed] [Google Scholar]
- 9.Kusano T, Yamaguchi K, Berberich T, Takahashi Y. The polyamine spermine rescues Arabidopsis from salinity and drought stresses. Plant Signal Behav. 2007;2:250–251. doi: 10.4161/psb.2.4.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta RA, Cassol T, Li N, Ali N, Handa AK, Matto AK. Engineered polyamine accumulation in tomato enhances phytonutrient content, juice quality and vine life. Nat Biotechnol. 2002;20:613–618. doi: 10.1038/nbt0602-613. [DOI] [PubMed] [Google Scholar]
- 11.Mattoo AK, Sobolev AP, Neelam A, Goyal RK, Handa AK, Segre AL. Nuclear magnetic resonance spectroscopy-based metabolite proWling of transgenic tomato fruit engineered to accumulate spermidine and spermine reveals enhanced anabolic and nitrogen-carbon interactions. Plant Physiol. 2006;142:1759–1770. doi: 10.1104/pp.106.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeHart GW, Jin T, McCloskey DE, Pegg AE, Sheppard D. The α9β1 integrin enhances cell migration by polyamine-mediated modulation of an inward-rectifier potassium channel. Proc Natl Acad Sci USA. 2008;105:7188–7193. doi: 10.1073/pnas.0708044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin-Tanguy Y. The occurrence and possible functions of hydroxycinnamoyl acid amines in plant. Plant Growth Regul. 2008;3:381–399. [Google Scholar]
- 14.Bagni N, Tassoni A. Biosynthesis, oxidation and conjugation of aliphatic polyamines in higher plants. Amino Acids. 2001;20:301–317. doi: 10.1007/s007260170046. [DOI] [PubMed] [Google Scholar]
- 15.Biondi S, Scaramagli S, Capitani F, Altamura MM, Torrigiani P. Methyl jasmonate upregulates biosynthetic gene expression, oxidation and conjugation of polyamines, and inhibits shoot formation in tobacco thin layers. J Exp Bot. 2001;52:231–242. [PubMed] [Google Scholar]
- 16.Creus JA, Eucuentra A, Gavalda EG, Barcelo J. Binding of polyamines to different macromolecules in plants. In: Galston AW, Tiburcio AF, editors. Polyamines as Modulators of Plant Development. Madrid: Ediciones Peninsular; 1991. pp. 30–34. [Google Scholar]
- 17.Flores H. Changes in polyamine metabolism in response to abiotic stress'. In: Slocum RD, Flores HE, editors. Biochemistry and Physiology of Polyamines in Plants. 1991. [Google Scholar]
- 18.Hawel L, Tjandrawinata RR, Fukumoto GH, Byus CV. Biosynthesis and selective export of 1,5-diaminopentane (cadaverine) in mycoplasma-free cultured mammalian cells. J Biol Chem. 1994;269:7412–7418. [PubMed] [Google Scholar]
- 19.Cvikrová M, Gemperlová L, Eder J, Zazimalová E. Excretion of polyamines in alfalfa and tobacco suspension-cultured cells and its possible role in maintenance of intracellular polyamine contents. Plant Cell Repts. 2008;27:1147–1156. doi: 10.1007/s00299-008-0538-5. [DOI] [PubMed] [Google Scholar]
- 20.Moschou PN, Paschalidis KA, Delis ID, Andriopoulou AH, Lagiotis GD, Yakoumakis DI, Roubelakis-Angelakis KA. Spermidine exodus and oxidation in the apoplast induced by abiotic stress is responsible for H2O2 signatures that direct tolerance responses in tobacco. Plant Cell. 2008;20:1708–1724. doi: 10.1105/tpc.108.059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanfrey C, Sommer S, Mayer MJ, Burtin D, Michael AJ. Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J. 2001;27:551–560. doi: 10.1046/j.1365-313x.2001.01100.x. [DOI] [PubMed] [Google Scholar]
- 22.Wu G, Bazer FW, Hu J, Hohnson GA, Spencer TE. Polyamine synthesis from proline in the developing porcine placenta. Biol Reprod. 2005;72:842–850. doi: 10.1095/biolreprod.104.036293. [DOI] [PubMed] [Google Scholar]
- 23.Minguet EG, Vera-Sirera F, Marina A, Carbonell J, Blazquez MA. Evolutionary diversification in polyamine biosynthesis. Mol Biol Evol. 2008;25:2119–2128. doi: 10.1093/molbev/msn161. [DOI] [PubMed] [Google Scholar]
- 24.Murray-Stewart T, Wang Y, Devereux W, Casero RA. Cloning and characterization of multiple human polyamine oxidase splice variants that code for isoenzymes with different biochemical characteristics. Biochem J. 2003;368:673–677. doi: 10.1042/BJ20021587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeller EA. Uber den enzymatischen Abbau von Histamin und Diaminen 2. Mitteilung. Helv Chim Acta. 1938;21:880–890. (Ger). (1938a) [Google Scholar]
- 26.Zeller EA. Zur Kenntnis der Diamin-oxydase 3. Mitteilung über den enzymatischen abbau von polyaminen. Helv Chim Acta. 1938;21:1645–1665. (Ger). [Google Scholar]
- 27.Bolenius FN, Seiler N. Acetylderivatives as intermediates in polyamine catabolism. Int J Biochem. 1981;13:287–292. doi: 10.1016/0020-711x(81)90080-x. [DOI] [PubMed] [Google Scholar]
- 28.Beard ME, Baker R, Conomos P, Pugatch D, Holtzman E. Oxidation of oxalate and polyamines by rat peroxisomes. J Histochem Cytochem. 1985;33:460–464. doi: 10.1177/33.5.3921604. [DOI] [PubMed] [Google Scholar]
- 29.Wu T, Yankovskaya V, McIntire WS. Cloning, sequencing and heterologous expression of the murine peroxisomal flavoprotein N1-acetylated polyamine oxidase. J Biol Chem. 2003;278:20514–20525. doi: 10.1074/jbc.M302149200. [DOI] [PubMed] [Google Scholar]
- 30.Tavladoraki P, Rossi MN, Saccuti G, Perez-Amador MA, Polticelli F, Angelini R, Ferderico R. Heterologous expression and biochemical characterization of a polyamine oxidase from Arabidopsis involved in polyamine back conversion. Plant Physiol. 2006;141:1519–1532. doi: 10.1104/pp.106.080911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moschou PN, Sanmartin M, Andriopoulou AH, Rojo E, Sanchez-Serrano JJ, Roubelakis-Angelakis KA. Bridging the gap between plant and mammalian polyamine catabolism: a novel peroxisomal polyamine oxidase responsible for a full back-conversion pathway in Arabidopsis. Plant Physiol. 2008;147:1845–1857. doi: 10.1104/pp.108.123802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylase mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Jiang D, Yang W, He Y, Amasino RA. Arabidopsis relatives of the human lysine-specific demethylase1 repress the expression of FWA and Flowering Locus C and thus promote the floral transition. Plant Cell. 2007;19:2975–2987. doi: 10.1105/tpc.107.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casero RA, Jr, Pegg AE. Spermidine/spermine N1-acetyltransferase—the turning point in polyamine metabolism. FASEB J. 1993;7:653–661. [PubMed] [Google Scholar]
- 35.Seiler N. Catabolism of polyamines. Amino Acids. 2004;26:217–233. doi: 10.1007/s00726-004-0070-z. [DOI] [PubMed] [Google Scholar]
- 36.Seiler N, Duranton B, Raul F. The polyamine oxidase inactivator MDL 72527. Prog Drug Res. 2002;59:1–40. doi: 10.1007/978-3-0348-8171-5_1. [DOI] [PubMed] [Google Scholar]
- 37.Cohen SS. A Guide to the Polyamines. New York: Oxford University Press; 1998. [Google Scholar]
- 38.Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 39.Thomas T, Thomas TJ. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell Mol Life Sci. 2001;58:244–258. doi: 10.1007/PL00000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urano K, Hobo T, Shinozaki K. Arabidopsis ADC genes involved in polyamine biosynthesis are essential for seed development. FEBS Lett. 2005;579:1557–1564. doi: 10.1016/j.febslet.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 41.Imai T, Matsuyama Y, Hanzawa T, Akiyama M, Tamaoki H, Saji Y, Shirano T, Kato H, Hayashi D, Shibata S, Tabata Y, Komeda Takahashi T. Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol. 2004;135:1565–1573. doi: 10.1104/pp.104.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imai A, Akiyama T, Kato T, Sato S, Tabata S, Yamamoto KT, Takahashi T. Spermine is not essential for survival of Arabidopsis. FEBS Lett. 2004:556. doi: 10.1016/s0014-5793(03)01395-4. [DOI] [PubMed] [Google Scholar]
- 43.Ge C, Cui X, Wang Y, Hu Y, Fu Z, Zhang D, Cheng Z, Li J. BUD2, encoding an S-adenosylmethionine decarboxylase, is required for Arabidopsis growth and development. Cell Res. 2006;16:446–456. doi: 10.1038/sj.cr.7310056. [DOI] [PubMed] [Google Scholar]
- 44.Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem (Tokyo) 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chattopadhyay MK, Myung HP, Tabor H. Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc Natl Acad Sci USA. 2008;105:6554–6559. doi: 10.1073/pnas.0710970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Kessler M, Ruiz OA, Maiale S, Ruiz-Herrera J, Jiménez-Bremont JF. Polyamine metabolism in maize tumors induced by Ustilago maydis. Plant Physiol Biochem. 2008;46:805–814. doi: 10.1016/j.plaphy.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Cona A, Rea G, Angelini R, Federico R, Tavladoraki P. Functions of amine oxidases in plant development and defense. Trends Plant Sci. 2006;11:80–88. doi: 10.1016/j.tplants.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 48.Paschalidis, Roubelakis-Angelakis Sites and regulation of polyamine catabolism in the tobacco plant. Correlations with cell division/expansion, cell cycle progression and vascular development. Plant Physiol. 2005;138:2174–2184. doi: 10.1104/pp.105.063941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cona A, Cenci F, Cervelli M, Federico R, Mariottini P, Moreno S, Angelini R. Polyamine Oxidase, a hydrogen peroxide-producing enzyme, is upregulated by right and downregulated by auxin in the outer tissues of the maize mesocotyl. Plant Physiol. 2003;131:803–813. doi: 10.1104/pp.011379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urano K, Yoshiba Y, Nanjo T, Igarashi Y, Seki M, Sekiguchi F, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant Cell Environ. 2003;26:1917–1926. [Google Scholar]
- 51.Walters DR. Polyamines and plant disease. Phytochemistry. 2003;64:97–107. doi: 10.1016/s0031-9422(03)00329-7. [DOI] [PubMed] [Google Scholar]
- 52.Walters DR. Resistance to plant pathogens: possible roles for free polyamines and polyamine catabolism. New Phytol. 2003;159:109–115. doi: 10.1046/j.1469-8137.2003.00802.x. [DOI] [PubMed] [Google Scholar]
- 53.Liu B, Sutton A, Sternglanz R. A yeast polyamine acetyltransferase. J Biol Chem. 2005;280:16659–16664. doi: 10.1074/jbc.M414008200. [DOI] [PubMed] [Google Scholar]
- 54.Chattopadhyay MK, Tabor CW, Tabor H. Absolute requirement of spermidine for growth and cell cycle progression of fission yeast (Schizosaccharomyces pombe) Proc Natl Acad Sci USA. 2002;99:10330–10334. doi: 10.1073/pnas.162362899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasukabe Y, he L, Nada K, Misawa S, Ihara I, Tachibana S. Overexpression of spermidine synthase enhances tolerance to multiple environmental stress and upregulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant Cell Physiol. 2004;45:712–722. doi: 10.1093/pcp/pch083. [DOI] [PubMed] [Google Scholar]
- 56.Kasukabe N, Watanabe-Sugimoto M, Matsuoka K, Okuma E, Obi I, Nakamura Y, Shimoishi Y, Murata Y, Kakutani T. Expression and Ca2+ dependency of plasma membrane K+ channels of tobacco suspension cells adapted to salt stress. Plant Cell Physiol. 2006;47:1674–1677. doi: 10.1093/pcp/pcl030. [DOI] [PubMed] [Google Scholar]
- 57.Capell T, Bassie L, Christou P. Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc Natl Acad Sci USA. 2004;101:9909–9914. doi: 10.1073/pnas.0306974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen XP, Pang XM, Matsuda N, Kita M, Inoue H, Hao YJ, Honda C, Moriguchi T. Overexpression of the apple spermidine synthase gene in pear confers multiple abiotic stress tolerance by altering polyamine titers. Transgenic Research. 2008;17:251–263. doi: 10.1007/s11248-007-9098-7. [DOI] [PubMed] [Google Scholar]
- 59.Urano K, Yoshiba Y, Nanjo T, Ito T, Yamaguchi-Shinozaki K, Shinozaki K. Arabidopsis stress-inducible gene for arginine decarboxylase AtADC2 is required for accumulation of putrescine in salt tolerance. Biochem Biophys Res Commun. 2004;313:369–375. doi: 10.1016/j.bbrc.2003.11.119. [DOI] [PubMed] [Google Scholar]
- 60.Moschou PN, Delis ID, Paschalidis KA, Roubelakis-Angelakis KA. Transgenic tobacco plants overexpressing polyamine oxidase are not able to cope with oxidative burst generated by abiotic factors. Physiol Plant. 2008;133:140–156. doi: 10.1111/j.1399-3054.2008.01049.x. [DOI] [PubMed] [Google Scholar]
- 61.Yamaguchi K, Takahashi Y, Berberich T, Imai A, Miyazaki A, Takahashi T, Michael A, Kusano T. The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett. 2006;580:6783–6788. doi: 10.1016/j.febslet.2006.10.078. [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi K, Takahashi Y, Berberich T, Imai A, Takahashi T, Michael A, Kusano T. A protective role for the polyamine spermine against drought stress in Arabidopsis. Biochem Biophys Res Commun. 2007;352:486–490. doi: 10.1016/j.bbrc.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 63.Kusano T, Yamaguchi K, Berberich T, Takahashi Y. Advances in polyamine research in 2007. J Plant Res. 2007;120:345–350. doi: 10.1007/s10265-007-0074-3. [DOI] [PubMed] [Google Scholar]
- 64.Uehara Y, Takahashi Y, Berberich T, Miyazak A, Takahashi H, Matsui K, Ohme-Takagi M, Satoh H, Terauchi R, Kusano T. Tobacco ZFT1, a transcriptional repressor with a Cys2/His2 type zinc finger motif that functions in spermine-signaling pathway. Plant Mol Biol. 2005;59:435–448. doi: 10.1007/s11103-005-0272-0. [DOI] [PubMed] [Google Scholar]
- 65.Tang W, Charles TM, Newton RJ. Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol Biol. 2005;59:603–617. doi: 10.1007/s11103-005-0451-z. [DOI] [PubMed] [Google Scholar]
- 66.Tang W, Newton RJ, Li C, Charles TM. Enhanced stress tolerance in transgenic pine expressing the pepper CaPF1 gene is associated with the polyamine biosynthesis. Plant Cell Rep. 2007;26:115–124. doi: 10.1007/s00299-006-0228-0. [DOI] [PubMed] [Google Scholar]
- 67.Tun NN, Santa-Catarina C, Begum T, Silveira V, Handro W, Floh EI, Scherer GF. Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 2006;47:346–354. doi: 10.1093/pcp/pci252. [DOI] [PubMed] [Google Scholar]
- 68.Yamasaki H, Shimoji H, Ohshiro Y, Sakihama Y. Inhibitory effects of nitric oxide on oxidative phosphorylation in plant mitochondria. Nitric Oxide. 2001;5:261–270. doi: 10.1006/niox.2001.0353. [DOI] [PubMed] [Google Scholar]
- 69.Romero-Puertas MC, Perazzolli M, Zago ED, Delledonne M. Nitric oxide signalling functions in plant-pathogen interactions. Cell Microbiol. 2004;6:795–803. doi: 10.1111/j.1462-5822.2004.00428.x. [DOI] [PubMed] [Google Scholar]
- 70.Yamasaki H, Cohen MF. NO signal at the crossroads: polyamine-induced nitric oxide synthesis in plants. Trends Plant Sci. 2006;11:522–524. doi: 10.1016/j.tplants.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 71.Kusano T, Berberich T, Tateda C, Takahashi Y. Polyamines: Essential factors for growth and survival. Planta. 2008;228:367–381. doi: 10.1007/s00425-008-0772-7. [DOI] [PubMed] [Google Scholar]
- 72.Tisi A, Angelini R, Cona A. Wound healing in plants: Cooperation of copper amine oxidase and flavin-containing polyamine oxidase. Plant Signal Behav. 2008;3:204–206. doi: 10.4161/psb.3.3.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoda H, Hiroi Y, Sano H. Polyamine oxidase is one of the key elements for oxidative burst to induce programmed cell death in tobacco cultured cells. Plant Physiol. 2006;142:193–206. doi: 10.1104/pp.106.080515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoda H, Yamaguchi Y, Sano H. Induction of hypersensitive response by hydrogen peroxide produced through polyamine degradation in tobacco plants. Plant Physiol. 2003;132:1973–1981. doi: 10.1104/pp.103.024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takahashi Y, Berberich T, Yamashita K, Uehara Y, Miyazaki A, Kusano T. Identification of tobacco HIN1 and two closely related genes as spermine-responsive genes and their differential expression during the Tobacco mosaic virus-induced hypersensitive response and during leaf- and flower-senescence. Plant Mol Biol. 2004;54:613–622. doi: 10.1023/B:PLAN.0000038276.95539.39. [DOI] [PubMed] [Google Scholar]
- 76.Kamada-Nobusada T, Hayashi M, Fukazawa M, Sakakibara H, Nishimura M. A putative peroxisomal polyamine oxidase, AtPAO4, is involved in polyamine catabolism in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1272–1282. doi: 10.1093/pcp/pcn114. [DOI] [PubMed] [Google Scholar]
- 77.Cervelli M, Di Caro O, Di Penta A, Angelini R, Federico R, Vitale A, Mariottini P. A novel C-terminal sequence from barley polyamine oxidase is a vacuolar sorting signal. Plant J. 2004;40:410–418. doi: 10.1111/j.1365-313X.2004.02221.x. [DOI] [PubMed] [Google Scholar]