Abstract
Heterotrimeric G-proteins are a class of signal transduction proteins highly conserved throughout evolution that serve as dynamic molecular switches regulating the intracellular communication initiated by extracellular signals including sensory information. This property is achieved by a guanine nucleotide cycle wherein the inactive, signaling-incompetent Gα subunit is normally bound to GDP; activation to signaling-competent Gα occurs through the exchange of GDP for GTP (typically catalyzed via seven-transmembrane domain G-protein coupled receptors [GPCRs]), which dissociates the Gβγ dimer from Gα-GTP and initiates signal transduction. The hydrolysis of GTP, greatly accelerated by “Regulator of G-protein Signaling” (RGS) proteins, returns Gα to its inactive GDP-bound form and terminates signaling. Through extensive characterization of mammalian Gα isoforms, the rate-limiting step in this cycle is currently considered to be the GDP/GTP exchange rate, which can be orders of magnitude slower than the GTP hydrolysis rate. However, we have recently demonstrated that, in Arabidopsis, the guanine nucleotide cycle appears to be limited by the rate of GTP hydrolysis rather than nucleotide exchange. This finding has important implications for the mechanism of sugar sensing in Arabidopsis. We also discuss these data on Arabidopsis G-protein nucleotide cycling in relation to recent reports of putative plant GPCRs and heterotrimeric G-protein effectors in Arabidopsis.
Key words: Arabidopsis, glucose, G-protein, nucleotide exchange, RGS protein
Canonical and Unusual GTPase Cycles
G-protein coupled receptors (GPCRs) normally serve as catalytic activators of heterotrimeric G-proteins (Gαβγ) by exchanging GTP for the bound GDP on the Gα subunit. This guanine nucleotide exchange factor (GEF) activity of GPCRs is the initial step in the mammalian G-protein cycle and the resultant separation of GTP-bound Gα from Gβγ determines the onset of various intracellular signaling pathways that govern critical physiological responses to extracellular cues. Heterotrimeric G-proteins also play a critical role in both cell-proliferation and glucose-signaling pathways in the model plant organism Arabidopsis thaliana, which has a single canonical Gα subunit (AtGPA1), one Gβ subunit, and two Gγ subunits.1 Currently for Arabidopsis, no bona fide GPCR has been described as possessing direct guanine nucleotide exchange factor (GEF) activity for the AtGPA1-containing heterotrimer1–4 (see also the subsequent section, ‘Do plants have GPCRs?’). However, the Arabidopsis RGS protein AtRGS1 is reminiscent of a GPCR in its protein domain architecture, consisting of seven transmembrane (TM)-spanning regions; AtRGS1 is unique in that its 7TM region is followed by an intracellular C-terminal RGS domain that acts in vitro as a GTPase-accelerating protein for AtGPA1 (a Gα “GAP”; also known as “GTPase-activating protein” in non-Gα-related contexts) and functions in cell proliferation and sugar-sensing pathways in vivo.5–8
Biochemistry of the Arabidopsis G-Protein Cycle
Despite its identification and cloning nearly 20 years ago,9 a biochemical characterization had not been carried out on the Arabidopsis thaliana G-protein α subunit AtGPA1. This contrasts with extensive in vitro enzymological characterizations of mammalian G-proteins.10 Although the rate constants for the guanine nucleotide cycle differ among Gα subunits, a commonality among all characterized mammalian Gα subunits is that the rate-limiting step of the G-protein cycle is nucleotide exchange and not GTP hydrolysis.10 This characteristic ensures that GPCR-mediated GEF activity is the singular determinant for signal onset. This brings up an interesting technical observation that is pertinent to much of the interpretations and hypotheses presented subsequently. The fact that the nucleotide exchange rate can be rate-limiting in the in vitro G-protein cycle makes it crucial that the nucleotide exchange rate (measured by quantifying the rate of GDP release or GTPγS binding) and the catalytic rate of GTP hydrolysis (measured by single turnover GTPase assay) are both measured.11 The facile assumption that catalytic and steady state GTPase rate constants are equivalent is almost never correct with heterotrimeric GTPases.11 This key consideration, although well-known to mammalian G-protein biochemists, has been generally overlooked by plant G-protein biochemists.
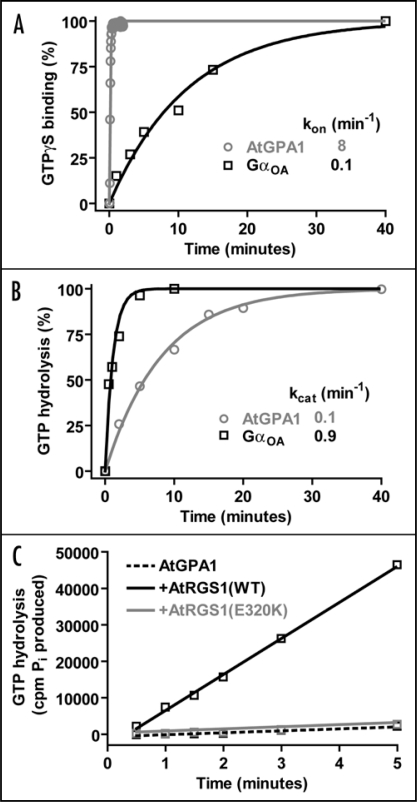
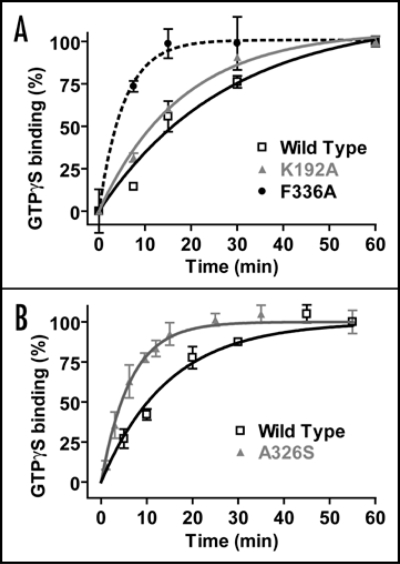
In a recent report, we described the biochemical characterization of AtGPA1 with respect to its guanine nucleotide cycle.6 In a surprising twist, AtGPA1 has a nucleotide cycle in which GTP hydrolysis, rather than nucleotide exchange, is the rate-limiting step.6 Enzymological analysis revealed a rate constant for spontaneous nucleotide exchange that is over 20-fold faster for AtGPA1 than for any known mammalian Gα subunit (Fig 1A). Moreover, the rate constant for GTP hydrolysis was found to be over 100-fold slower than this extremely rapid nucleotide exchange rate (Fig. 1A and B).6 The RGS domain of AtRGS1 was found to potently and robustly stimulate the relatively slow GTP hydrolysis rate of AtGPA1 (Fig 1C); moreover, the RGS domain-accelerated GTP hydrolysis rate at steady-state was seen to approximate the single turnover GTP hydrolysis rate.6 All told, our analysis predicts that AtGPA1 is likely 99% bound to GTP at steady-state, which differs significantly from mammalian Gαo (the next-fastest spontaneous exchanger currently known) that is predicted to be only 10% GTP-loaded at steady state.6 One obvious caveat to these results is the finding from mammalian heterotrimeric G-protein studies that, under certain conditions, Gβγ subunits can dampen the spontaneous nucleotide exchange rate of Gα·GDP.12,13 This process is known as guanine nucleotide dissociation inhibitor (GDI) activity, and is thought to be a mechanism by which spontaneous G-protein activity is prevented in vivo. However, the largest reported magnitude of the GDI effect exerted by Gβγ is 5-fold,12 and it would require approximately a 3-order of magnitude larger GDI effect to make nucleotide release the in vitro rate limiting step for the Arabidopsis AtGPA1.6 This is obviously an issue that needs to be resolved but, for this to happen, it will require reconstitution of the Arabidopsis heterotrimer with full post-translational lipid modifications of the Gα and Gγ moieties.14 Moreover, although the biochemical role of GDI activity has been demonstrated in vitro, it has not been shown in vivo,12–14 even in the context of rigorously characterized mammalian signal transduction paradigms. To achieve such an in vivo demonstration, a mutation which inhibits Gβγ-mediated GDI activity, but not other properties of the heterotrimer, would need to be identified.
Figure 1.
Enzymological properties of the Arabidopsis heterotrimeric G-protein alpha subunit (AtGPA1) in comparison to human GαoA. (A) GTPγS binding rates of AtGPA1 and GαoA were measured using standard methods.6,20 Data were fit using a single exponential function and rate constants are presented in min−1. Note: the observed GTPγS binding rate (kon) is equivalent to the rate of GDP release, as GDP release is the rate limiting step in this process.11 (B) The GTP hydrolysis rates of AtGPA1 and GαoA were measured using single turnover GTP hydrolysis78 (i.e., using Gα subunits preloaded with [γ-32P]GTP). Data were fit using a single exponential function and rate constants are presented in min−1. (C) Steady state GTP hydrolysis of 100 nM AtGPA1 was measured using [γ-32P]GTP, as described.6 AtGPA1 was incubated with either buffer (dotted black line), 250 nM wild-type AtRGS1(black line), or 250 nM E320K AtRGS1(grey line). The latter protein serves as a negative control for this experiment as it harbors a glutamate-320 to lysine (E320K) charge-reversal mutation that cripples RGS domain-mediated GAP activity, as previously described.6,79 A general phenomenon of RGS proteins is that steady state GTPase acceleration cannot be observed in the absence of a GEF80 as GDP release (not GTP hydrolysis) is rate limiting step in the G-protein cycle.11 However, in this case, the steady state GTPase rate of AtGPA1 is greatly accelerated in the presence of AtRGS1-mediated GAP activity, highlighting the fact that GDP release is not rate-limiting for this plant Gα subunit. Observed rate constants were calculated by linear regression: AtGPA1 532 cpm/min, + wild-type AtRGS1 9867 cpm/min, + E320K AtRGS1 579 cpm/min.
Evolutionary Implications of the Arabidopsis G-Protein Cycle
The only other plant G-protein alpha subunit that has been analyzed biochemically is the rice Gα subunit (RGA1), which appears to have mammalian-like biochemical properties.15,16 RGA1 was described as having a nucleotide exchange rate of 0.014 min−1 (compare to ≈ 10 min−1 for AtGPA1) and a steady state GTP hydrolysis rate of 0.0075 min−1 (compare to ≈ 0.1 min−1 for AtGPA1). This is not surprising as significant sequence and predicted functional divergence between the Gα subunits of the dicotyledon Arabidopsis (AtGPA1) and the monocotyledon rice Oryza sativa (RGA1) has been described.1 Thus, it may be that rice has a G-protein cycle more akin to that of other species containing conventional GPCR GEFs and RGS protein GAPs. However, the RGA1 nucleotide exchange kinetic data reported by Seo et al. were obtained using only a relatively small excess of nucleotide to RGA1 (assays were described as containing 500 ng of RGA1 and 200 nM GTPγS in a final volume of 200 µl; this equates to 50 nM RGA1 being used with a 4-fold excess of GTPγS [200 nM]). It would only require a 3-fold change in either of the two RGA1 rate constants to make GTP hydrolysis the rate limiting step. Interestingly, Iwasaki and colleagues have also analyzed the kinetics of RGA1 GTP binding and hydrolysis.17 They found that the rates of GTP binding and hydrolysis were similar (kon = 0.36 min−1 and k=cat 0.44 min−1 at 20°C). Thus, based on their data, GTP hydrolysis is the rate limiting step in the rice G-protein cycle. However, it must be noted that the differences between the rate constant data of Iwasaki17 and Seo15 are 20- to 60-fold. The fact that the biochemical data of Iwasaki17 and Seo15 are so discordant makes it difficult to draw confident conclusions about the biochemistry of rice G-protein signaling.
Although our data with AtGPA1 appear to obviate the need for GPCR-mediated GEF activity in vivo, this does not explicitly discount a role for GEFs/GPCRs in Arabidopsis. Arabidopsis and several other plant species contain multiple ‘extra-large’ G-protein alpha subunits (XLGs) (refs. 18 and 19, and Willard FS, unpublished data). These proteins contain the all-helical and Ras-like domains common to Gα subunits, but also N-terminal extensions of unknown function. There is some evidence that the three AtXLGs redundantly regulate root development and sensitivity to sugars and abscisic acid.18 Intriguingly, these proteins lack a myristoylation consensus sequence typical of canonical Gα subunits, and GFP fusions of these proteins localize to the nucleus, rather than the plasma membrane, of plant cells.18 Furthermore, it remains to be determined whether AtXLGs are functional GTPases. One report claims to show GTP binding activity by AtXLG1;19 however, [35S] GTPγS binding was measured by autoradiography following SDS-PAGE. As GTP binding is non-covalent it is highly unlikely that [35S] GTPγS would stay bound during denaturing gel electrophoresis. It is well accepted that GTP binding in this context should only be measured using radioligand binding or fluorescence-based assays.6,13,20
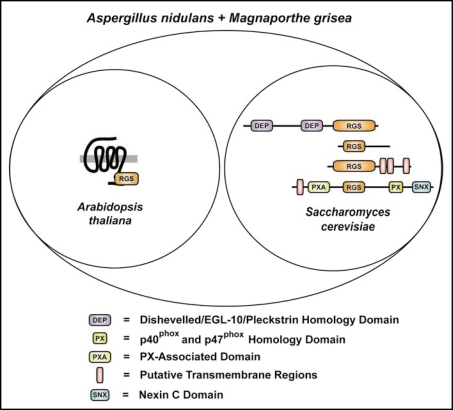
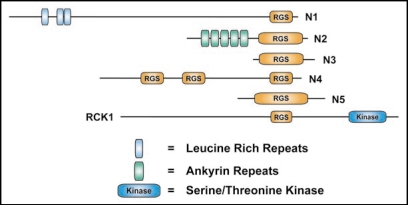
It is noteworthy that eukaryotes exhibit a diverse repertoire of RGS protein usage. Yeast typically possess four distinct RGS proteins with differing domain architectures (Fig 2). In the case of S. cerevisiae, this set is composed of the dual DEP domain-containing SST2, the isolated RGS domain protein RGS2, the predicted three transmembrane RGS protein RAX1, and the sorting nexin RGS protein MDM1. However, Aspergillus nidulans,21 Magnaporthe grisea,6,21 and several other fungi6 appear to contain these four canonical RGS proteins of yeast, as well as one or more AtRGS1-orthologous proteins (Fig 2). We also observed that the protozoan Trichomonas vaginalis has multiple AtRGS1-orthologous proteins,6 but we have not yet identified any canonical RGS proteins in this organism. These results therefore suggest that different eukaryotes employ distinctly different G-protein signaling mechanisms, involving the use of both 7TM GAPs and 7TM GEFs as exemplified by some fungal genomes. These data also suggest that there may be distinct differences between monocotyledon and dicotyledon plants in their G-protein signal transduction pathways. Further comparative functional genomic and physiological studies are needed to clarify these points; however, interesting observations can be noted from comparative genomic analyses of RGS proteins. For instance, the amoeba Dictyostelium discoideum is predicted to have a wide variety of RGS-domain containing proteins with novel domain structures (Fig. 3). Assuming these are not sequence annotation artifacts, these uncharacterized classes of RGS proteins provide an exciting window into the evolution of eukaryotic G-protein signaling systems. For example, the Dictyostelium RGS protein RCK1 (Fig. 3 and ref. 22) may be the ancestral homolog of G-protein coupled receptor kinases, which contain both kinase and RGS domains.23
Figure 2.
Selected RGS proteins of eukaryotic model organisms. Venn diagram illustrating the structural classes of RGS proteins identified in eukaryotic model organisms. Protein architecture schematics highlight the multiple domains found in the RGS proteins of Arabidopsis thaliana, Saccharomyces cerevisiae, Aspergillus nidulans and Magnaporthe grisea. Indicated are the archetypal RGS proteins of Arabidopsis (AtRGS1; GenBank accession, NP_189238) and S. cerevisiae (top to bottom: SST2; SwissProt accession P11972), Rgs2 (GenBank accession NP_014750), Rax1 (GenBank accession NP_014945), and yeast Mdm1 (GenBank accession NP_013603). Domain abbreviations: DEP, Dishevelled/EGL-10/pleckstrin homology domain; RGS, regulator of G-protein signaling domain; PXA, PX-associated domain; PX, p40phox and p47phox homology domain; SNX, Nexin C domain. Putative transmembrane regions are denoted by pink vertical bars.
Figure 3.
The predicted RGS proteins of Dictyostelium discodium. Schematic of the multiple domain-containing RGS proteins of Dictyostelium discoideum (GenBank accession numbers in parentheses): N1 (Q556I3), N2 (Q54MA7), N3 (Q54XJ6), N4 (Q54LD1), N5 (Q54M81), and RCK1 (XP_64197822). RGS, regulator of G-protein signaling domain; Kinase, serine/threonine kinase. Leucine rich repeats are denoted by blue vertical bars. Ankyrin repeats are denoted by green vertical bars. Note: many of the predicted Dictyostelium discoideum proteins contain polyasparagine tracts typical of this genome.81
Insights into the Molecular Basis of Rapid Guanine Nucleotide Exchange
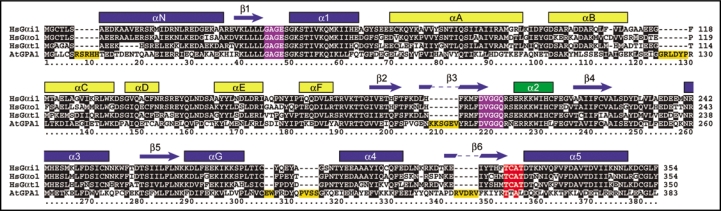
A multiple sequence alignment between AtGPA1 and mammalian Gαi/o subfamily members shows an overall high degree of amino acid conservation between these proteins; however, several notable insertions within the longer AtGPA1 primary sequence are apparent (Fig. 4). For example, six polypeptide insertions exist within AtGPA1 (Fig. 4), with one extending the N-terminus (7RSRHH11), one extending the αB/αC loop within the all-helical domain (124GRLDYP129), and four within the Ras-like GTPase fold (208KKSGEV213, 305EW306, 312PVSS315 and 343RVDRV347).
Figure 4.
Multiple sequence alignment of AtGPA1 and human (Hs) Gαi1, GαoA and rod transducin-Gαt1. Known secondary structure of Gα subunits (α-helices as bars; β-strands as arrows) is annotated within the Ras-like domain (blue), all-helical domain (yellow), and switch regions (green). Note: several polypeptide insertions are unique to AtGPA1 (orange boxes). In addition, the canonical TCAT motif within the β6/α5 loop (red box) differs within AtGPA1, although guanine base and phosphate contact positions (GAGE and DVGGQ motifs; purple boxes) are completely conserved. Accessions numbers for protein sequences: AtGPA1 (P18064), Gαi1 (P63096), GαoA (P09471) and Gαi1 (P11488).
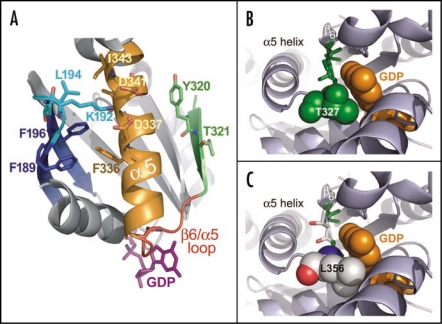
One of these inserts unique to AtGPA1 (amino acids 208KKSGEV213) occurs near the β2/β3 loop. In the crystallographic structures of mammalian Gα subunits, this loop makes several contacts with the C-terminal region of the α5 helix, and evidence suggests these contacts, thought to be disrupted by binding to activated receptors, are critical for restricting basal nucleotide exchange.24–26 Thus, an insertion near the AtGPA1 β2/β3 loop may result in disruption of α5 helix interactions normally found in mammalian Gα subunits, resulting in enhanced spontaneous nucleotide exchange of AtGPA1. Single amino acid changes in the β2/β3 loop are also known to modulate the kinetics of GDP release. The Gα-transducin β2/β3 loop residue K188, when mutated to alanine, engenders a 6-fold increase in nucleotide exchange.26 Mutation of this residue is predicted to disrupt a salt-bridge that involves two α5 helix aspartate residues and stabilizes the conformation of the α5 helix. As an independent demonstration of the importance of this salt-bridge, mutation of K192 to alanine in Gαi1 modestly increases the GDP release rate of Gαi1 (Fig. 5A). Similarly, mutation of other amino acid residues (such as F189, K192, L194, F196), which stabilize the conformation of the α5 helix, also increase spontaneous GDP release (as depicted in Fig. 6A).26 In a reciprocal manner, disruption of α5 helix interactions with β2/β3 regions can also modulate GDP release. For instance, Phe336, when mutated to alanine in Gα-transducin, results in a >160-fold increase in nucleotide exchange.26 Mutation to alanine is predicted to disrupt several hydrophobic interactions with the β2/β3 region (e.g., residues F189 and F196) thought to stabilize the ground-state or “resting” orientation of the α5 helix (Fig. 6A). As an independent demonstration of this interaction, we present data from Gαi1 showing that mutation of F336 to alanine results in a marked enhancement of spontaneous GDP release (Fig. 5A). Thus, we conclude that residues in the β2/β3 region, by virtue of modulating α5 helix stability and vice versa, may also be responsible for the rapid nucleotide exchange kinetics of AtGPA1.
Figure 5.
Effect of mutations in the β2/β3 loop, α5 helix, and the β6/α5 loop on the GDP release rate of Gαi1. The GDP release rate of wild type and mutant Gαi1 subunit was measured using [35S]GTPγS radioligand binding, as described.78 GTPγS binding is an accurate method of measuring GDP release, given that GDP release is the rate limiting step in the nucleotide exchange process.11 Data were fit to a single exponential function using GraphPad PRISM 3.0. Observed rate constants: (A) wild type, 0.036 min−1; K192A, 0.056 min−1; F336A, 0.1844 min−1 (B) wild type, 0.068 min−1; 0.16 min−1, A326S. Note: the rate enhancement engendered by the A326S mutation in this experiment was only 3-fold, not the 20-fold previously reported.32 We observed faster GDP release by A326S Gαi1 in some experiments. We hypothesize that this may be due to the idiosyncratic effects of polyoxyethylene 10-lauryl ether (lubrol) on GDP release, as described.12
Figure 6.
Structural analyses of the α5 helix and the β6/α5 loop in the regulation of GDP release. (A) The β6/α5 loop (red) plays a key role in regulating the binding and release of GDP (magenta sticks). The β6 strand (green) and α5 helix (orange) are each thought to regulate the disposition of this loop. Additionally, the β2/β3 loop (cyan) connecting the β2 and β3 strands (blue) is thought to indirectly affect GDP release by stabilizing the conformation of the α5 helix. Several residues governing this interaction in the β2/β3 loop and α5 helix are shown as sticks. Residues Y320 and T321 in the β6 strand may also stabilize the basal conformation of the α5 helix. Receptor-mediated disruption of these regions may ultimately induce a conformational change in the β6/α5 loop resulting in the release of GDP (reviewed in ref. 82). The structural representation was generated from PDB file: 1BOF.83 (B) Structural representation of residue T327 in Gαi1 highlights the role of β6/α5 loop in nucleotide exchange. Structure of wild type Gαi1 (PDB code: 1BOF)83 illustrates the proximity of the β6/α5 loop to the bound molecule of GDP (orange spheres). The highly conserved TCAT motif (green sticks) within the β6/α5 loop positions the T327 side chain (spheres) directly towards the GDP purine ring. This side chain perfectly accommodates the bound GDP molecule, making several potentially stabilizing contacts (not depicted). (C) AtGPA1 contains a unique TTAL motif within its β6/α5 loop. A modeled mutation of the TCAT motif within the Gαi1 structure to TTAL as present in AtGPA1 (green and white sticks) shows that the leucine side chain (spheres; numbered as in AtGPA1) introduces a likely steric clash with the GDP molecule. This unproductive orientation of the β6/α5 loop is predicted to reduce the affinity for GDP binding and result in a faster nucleotide exchange rate for AtGPA1. The AtGPA1 structural model was generated using the ‘mutagenesis’ function in PyMol (Delano Scientific; San Carlo, CA, USA). The structure of Gαi1 (PDB id 1BOF) was used as the basis for this model and the indicated residues were mutated in silico to corresponding residues in AtGPA1.
A second notable insertion in AtGPA1 (343RVDRV347; Fig 4) occurs near the α4/β6 loop. This region of Gα has long been considered a site for receptor/Gα interaction critical to G-protein activation.27–29 We have recently confirmed the α4/β6 loop as a critical determinant of Gαi1 interaction with, and agonist-evoked activation by, the D2-dopamine receptor.30 Insertion within this α4/β6 loop region of AtGPA1 serves as another attractive hypothesis for conformational differences leading to its enhanced nucleotide exchange rate.
The conserved Thr-Cys-Ala-Thr motif within the β6/α5 loop directly contacts GDP and contributes to the spontaneous nucleotide exchange rate (Fig. 4).31–33 The canonical amino acid sequence of the β6/α5 loop is TCAT whereas, in AtGPA1, it is TTAL. These two alterations within this loop most likely weaken interactions with the purine base of GDP (Fig. 6B). Mutation in the β6/α5 loop of Gαs(A366S) has been described in a human clinical population manifesting in testotoxicosis and pseudohypoparathyroidism.31 A key element of the mechanism of action of this mutation is a 20-fold increase in the rate of spontaneous GDP release.31 Mutation of the analogous residue in Gαi1 (A326S) has also been shown to increase GDP release by up to 25-fold.32 We have observed analogous results (Fig. 5B), although with less fold-enhancement of GDP release. The structure of Gαi1 (A326S) has been determined,32 and indeed interactions between the G-protein and the guanine nucleotide are altered, including loss of some contacts between the β6/α5 loop and the guanine base, and creation of new contacts with the nucleotide and the switch-I residue Arg178 and residues in the P-loop.32
A definitive determination of the mechanistic basis of weak GDP affinity by AtGPA1 may require high-resolution knowledge of the structural determinants of AtGPA1 bound to nucleotide. Modeling the β6/α5 loop of AtGPA1 onto the crystal structure of Gαi1 reveals that a T327L mutation (corresponding to Leu356 normally found in AtGPA1) creates a potentially unproductive steric clash between the hydrophobic leucine side chain and the guanine ring of GDP. Moreover, the OG1 oxygen of the T327 side chain within native Gαi1, which perfectly accommodates the GDP molecule, makes a stabilizing bond with the N1 nitrogen of the purine ring of GDP (Fig. 6B, not highlighted). The steric clash with GDP that likely results from the larger side chain of Leu356 within AtGPA1 could theoretically reduce the affinity of GDP and manifest into the higher spontaneous nucleotide exchange rate observed experimentally. While other segments of the AtGPA1 sequence could also contribute to the overall mechanism of enhanced GDP release, the TTAL sequence of the β6/α5 loop, a region implicated in receptor-mediated GDP release, most likely plays a key role.
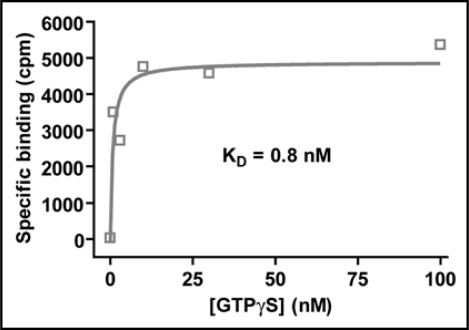
We have attempted to measure the affinity of AtGPA1 for GDP, but have been unsuccessful due to the lack of a high specific activity radioligand and the apparent low affinity of AtGPA1 for GDP (Willard FS, unpublished data). We have, however, been able to estimate the affinity range of AtGPA1 for Mg2+·GTPγS and, as in the case of mammalian Gα subunits, this is very tight. For instance, using 1 nM of AtGPA1, a KD value of 2 nM was obtained (Fig. 7); similarly, with 5 nM of AtGPA1, a KD value of 5 nM was obtained (Willard FS, unpublished data). These data indicate that the affinity of AtGPA1 for GTPγS is likely sub-nanomolar. The fact that AtGPA1 has a high affinity for GTPγS could be mistakenly interpreted as casting doubt on our data demonstrating that AtGPA1 has weak affinity for GDP. This, however, would be a fallacious argument. Heterotrimeric G-proteins are known to have an extremely high affinity for the non-physiological, non-hydrolyzable ligand Mg2+·GTPγS. This affinity is substantially higher than that of Mg2+·GTP, which in turn is much higher than that for GDP.13,32 Thus, rate constants are typically not quantifiable when GTPγS dissociation is measured for heterotrimeric G-proteins.13,32 In the presence of millimolar amounts of Mg2+, GTPγS binding is observed to be essentially irreversible over the stability time-course of the G-protein alpha subunit.13
Figure 7.
Saturation binding analysis of the affinity of Mg2+·GTPγS for AtGPA1. 1 nM AtGPA1 was mixed with various concentrations of [35S] GTPγS in the presence of 25 mM MgCl2.6,78 Bound GTPγS was quantified by filtration and liquid scintillation as described.6,78 Non-specific binding was determined in the presence of 100 mM unlabeled GTPγS. Specific binding was fit to a saturation binding isotherm (Y = Bmax + X / (KD + X)) using GraphPad PRISM 3.0.
The Physiology of Glucose Sensing by AtRGS1/AtGPA1
Our initial cloning and characterization of AtRGS1 suggested a unique topology of a 7TM ‘GPCR-like’ conformation with an RGS domain residing at the intracellular C-terminus.5,34,35 However, the biochemical function of AtRGS1 (e.g., GEF and/or GAP activities) remains elusive. Our recent characterization of the AtGPA1 guanine nucleotide cycle portends a critical function of RGS domain-mediated GAP activity in proper in vivo signaling. Phenotypic analyses of Arabidopsis lines expressing a loss-of-function point mutant of AtRGS1 (E320K, a charge reversal on the predicted Gα interaction surface of the RGS domain) confirmed the importance of RGS domain-mediated GAP activity to proper sugar signaling.6 Whereas expression of wild-type AtRGS1 results in reduced hypocotyl length, AtRGS1(E320K) mutant lines display normal hypocotyl lengths. Additionally, AtRGS1(E320K) was unable to rescue AtRGS1-null allele effects on glucose-mediated growth arrest and also did not exhibit the glucose-hypersensitive phenotype commonly seen with plants overexpressing wild-type AtRGS1.6 Together with our biochemical analysis of AtGPA1, these results highlight that the GAP activity of AtRGS1 is a critical determinant of Arabidopsis response to environmental glucose, consistent with the hypothesis that AtRGS1 is a glucose-regulated GAP for constitutively GTP-bound AtGPA1. However, the role for heterotrimeric G-proteins in Arabidopsis signal transduction is multifaceted. The Arabidopsis G-protein pathway is a crucial regulator of cell proliferation1,36 and abscisic acid-induced stomatal opening.37 Furthermore, the Arabidopsis G-protein pathway is also implicated in seed germination and the unfolded protein response.8,38,39
Sugar Sensing GPCR Systems in Eukaryotes
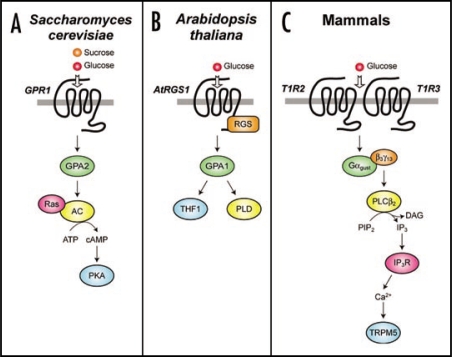
Nutrient detection is an important cellular function and, in eukaryotes, multiple independent signal transduction pathways have evolved for this critical aspect of environmental sensing. However, several of these pathways have a commonality in being mediated by heterotrimeric G-protein linked systems (Fig. 8). Sugar sensing in plants is convoluted, and it appears that multiple pathways are operant.40 However, the G-protein linked pathway appears to couple extracellular glucose sensing to the regulation of developmental processes via AtRGS1,5–7 AtGPA1,41 and possibly the AtGPA1 interacting plastid protein THF1.41 Glucose stimulates interactions between AtRGS1 and AtGPA1,6 between AtGPA16 and THF1,41 and probably between AtGPA1 and as yet undefined effector systems to regulate cellular physiology. Analogous to this Arabidopsis pathway, S. cerevisiae detects glucose or sucrose via the GPCR GPR1.42 This receptor has a millimolar affinity for sugars, and activates a signaling pathway via the heterotrimeric G-protein GPA2, the monomeric G-protein Ras, the generation of cyclic AMP, and the resultant activation of PKA.43 This pathway is involved in nutrient-regulated cell growth and stress response. In mammals, a functional glucose-sensing heterodimeric receptor has been described44–46 that is formed by the GPCRs T1R2 and T1R3. The T1R2/3 heterodimer is present on taste cells and detects ingested sugars. This receptor heterodimer is thought to primarily signal through a G-protein heterotrimer of Gα gustducin/Gβ3/Gγ13 to activate phospholipase Cβ2 (PLCβ2) and he TRPM5 channel.44,47 Peripheral neuronal signals are generated by this signaling pathway to form the sensation of taste.44,47 Recent studies have suggested further roles for this conserved signaling cassette in the detection of nutrients. The T1R2/3 heterodimer, gustducin, PLCβ2, and TRPM5 are present in the enteroendocrine cells of the gut.48,49 Mice lacking gustducin are deficient in glucose-induced GLP secretion.48,49 Thus, GPCRs mediate a wide variety of glucose-dependent physiological responses in multiple organisms, and appear to be evolutionarily critical in this regard.
Figure 8.
Comparison of GPCR-based sugar sensing in Saccharomyces cerevisiae, Arabidopsis thaliana and mammals. (A) A model of sugar sensing in Saccharomyces cerevisiae.40,84 Binding of glucose and sucrose to GPR1 leads to activation of the Gα subunit GPA2. Activated Ras and activated GPA2 bind independently to adenylate cyclase and stimulate its production of cyclic AMP (cAMP). This second messenger then promotes activation of the protein kinase A (PKA) tetramer by binding to the regulatory subunits and promoting dissociation of the complex. (B) A model of sugar sensing in Arabidopsis thaliana. Binding of glucose to AtRGS1 leads to regulation of the Gα subunit AtGPA1. GPA1 can regulate the plastid protein thylakoid formation 1 (THF1) 41 and phospholipase D (PLD).71 (C) A model for sugar sensing in mammals.47,78 Binding of glucose to type 1 taste GPCRs (T1R) 2 and 3 leads to dissociation of the Gα subunit gustducin (Gαgust) from the Gβ3γ13 dimer. Gβ3γ13 then activates phospholipase Cβ2 (PLCβ2), which in turn hydrolyzes phosphatidylinositol bisphosphate (PIP2) into diacylglycerol (DAG) and inositol trisphosphate (IP3). The latter second messenger activates IP3 receptors (IP3R), which release intracellular Ca2+ from privileged cellular stores (such as the ER). Intracellular Ca2+ then opens transient receptor potential cation channel, subfamily M, member 5 (TRPM5), which leads to an influx of Na+ and depolarization of the cell.
Do Plants have G-Protein Coupled Receptors?
As previously described, the first plant heterotrimeric G-protein was cloned in 1990—a timing essentially coincident with the initial cloning of Gα subunits from many mammalian species and other model organisms.9,50 As of 2008, however, no plant GPCR has been definitively demonstrated to exist, whereas numerous physiological and biochemical proofs of GPCR activity exist in mammals, yeast, flies, fungi and worms.34,51,52 Our biochemical data suggest that a canonical GPCR which catalyzes guanine nucleotide exchange on AtGPA1·GDP/Gβγ is unlikely to exist; however, this does not deny the possibility of the existence of other membrane receptors that can modulate G-protein activity either indirectly or directly. Glucose-modulated GAP activity of the AtRGS1 receptor would be a pertinent example of this, for instance.6
A naïve participant in the plant G-protein field should be very circumspect in their analysis of reports purporting to demonstrate GPCR-like activity by plant proteins, either in vitro or in vivo. The defining properties of GPCRs are typically two-fold: (1) they are seven transmembrane spanning (7TM), integral membrane proteins, and (2) they activate heterotrimeric G-proteins by binding to Ga·GDP/Gβγ and catalyzing the release of GDP. Thus, 7TM proteins which are physiologically implicated in the G-protein pathway are not necessarily GPCRs. There are numerous predicted 7TM proteins in plants.4 Recently, the Arabidopsis protein GCR2 (G-protein coupled receptor 2) was described as a specific GPCR for the plant hormone abscisic acid.53
Identification of GCR2 as a GPCR was based on four lines of evidence. Firstly, GCR2 was predicted to be a 7TM protein using bioinformatics.53 However, this data was obtained using algorithms with 70–90% false positive prediction rates.54 Moreover, it is apparent that the authors ‘cherry-picked’ the derived transmembrane domain prediction data to support their hypothesis of 7TM domains being present in GCR2.55 Two independent reports have refuted the 7TM topology of GCR2.3,56 Moreover, we have described that GCR2 is most likely a plant homolog of cytoplasmic lanthionine synthetases56 based on sequence homology57 and protein fold recognition.58 Secondly, the signaling-defective phenotype of GCR2-deficient plants was reported to support GCR2 as being responsible for all effects of the hormone abscisic acid (ABA) in Arabidopsis.53 However, this data could not be repeated by other investigators.2 Thirdly, GCR2 was shown to interact with AtGPA1 using a variety of techniques.53,55 Fourthly, GCR2 was shown to be a stereospecific receptor for abscisic acid.53 These third and fourth points have yet to be reproduced by other researchers. Based on the totality of the original data presented, and its refutation subsequent to its first publication, we conclude that GCR2 is not a GPCR, nor a transmembrane protein, nor a likely cellular receptor for ABA-induced physiological responses.2,3 However, it is possible that GCR2 is involved in abscisic acid metabolism or synthesis, and this would account for its observed high affinity binding interactions,53 and the apparent idiosyncrasies involved in reproducing the physiological experiments.2
The best GPCR candidate in plants thus far has been the Arabidopsis protein GCR1 (G-protein coupled receptor 1). GCR1 is a bona fide 7TM protein, with appreciable similarity to the Dictyostelium cAMP receptor.59 GCR1 has been shown to interact with AtGPA1, and GCR1-deficient plants are hypersensitive to abscisic acid.59 However, the role of GCR1 in the Arabidopsis G-protein signaling pathway is not clear-cut, as GCR1 appears to have multiple functions independent of heterotrimeric G-proteins.60 Certainly, the data demonstrating physical interaction between AtGPA1 and GCR1 are not definitive.59 Co-immunoprecipitations using in vitro translated AtGPA1 and GCR1 were used to demonstrate interaction.59 It is difficult to comprehend how a 7TM protein such as GCR1 can be produced in a functional form using in vitro translation in the absence of a lipid membrane. The authors used a rabbit reticulocyte lysate based system (Novagen STP3) which would most likely need exogenous lipids or microsomal membranes to allow functional transmembrane protein production.61,62 Similarly, splitubiquitin complementation assays were used to show interaction between AtGPA1 and GCR1 in yeast.59 Interaction was observed between AtGPA1 fused to the C-terminal half of ubiquitin and the N-terminus of GCR1 fused to the N-terminal half of ubiquitin.59 These results are hard to understand from a topological standpoint, given that the N-terminus of GCR1 is predicted to be extracellular59 yet AtGPA1 is an intracellular protein. Thus, at the current juncture, there does not seem to exist sufficient, compelling data to demonstrate that a canonical GPCR exists in plants.
G-Protein Effectors in Plants
Another unresolved question in plants relates to the mechanisms by which G-protein signal transduction is mediated directly proximal to the G-protein. Effector systems in mammals have been well characterized biochemically, structurally, and in in vivo settings. However, little is known about Plantae G-protein effector systems. Generation of AtGPA1-null alleles in Arabidopsis allowed the analysis of the role of the G-protein pathways in stomatal opening.37 Abscisic acid inhibits guard cell K+ channels and, consequently, stomatal opening. ABA sensitivity is lost in AtGPA1-null plants, suggesting that K+ channels could be direct effectors of AtGPA1 or Arabidopsis Gβγ subunits, similar to how these channels serve as effectors within mammalian G-protein signaling pathways.37,63 However, ABA inhibition of K+ channels is thought to be indirect, as ABA appears to activate sphingosine kinase to produce sphingosine 1-phosphate, a signaling molecule considered to act upstream of AtGPA1.64 Thus, it remains to be determined how the Arabidopsis G-protein pathway directly regulates ion channels. The phospholipase D (PLD) enzymes have long been implicated as possible effectors of AtGPA1, based on analogy with the mammalian usage of phospholipase C (PLC) systems.65,66 However, some of the initial studies on PLD need to be reinterpreted based on our accumulated knowledge of Arabidopsis G-protein biochemisty. For instance, the wasp venom peptide mastoparan is frequently used as an ‘activator’ of plant G-protein pathways and, in particular, of PLC and PLD systems.67 Based on the unique nucleotide biochemistry of AtGPA1 as described above, mastoparan should not appreciably stimulate guanine nucleotide exchange by AtGPA1 as this is not a physiologically rate-limiting step.6 Thus, the use of mastoparan to demonstrate involvement of heterotrimeric G-proteins in Arabidopsis physiological processes must be reinterpreted. Indeed, this point has been demonstrated genetically in terms of mastoparan-activated MAPK activity that is independent of G-protein signaling.68 However, this does not discount the possibility that mastoparan may still bind to Gα and thereby elicit protein-protein interactions and/or GTPase activity. Most likely though, the effects of mastoparan on cellular systems in plants is mediated via other mechanisms, given that mastoparan has multiple cellular targets.69 Similarly, the use of bacterial toxins such as pertussis and cholera toxins may be misguided as applied to plant signaling systems. For example, pertussis toxin catalyzes ADP-ribosylation on a conserved C-terminal cysteine residue of Gαi subunits, yet this cysteine residue is not present in plant G-proteins.9,70
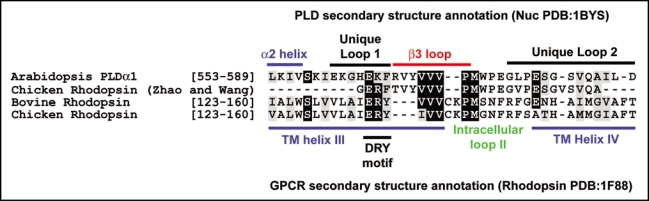
Wang and colleagues have investigated PLDα1 as a putative AtGPA1 effector.71 They demonstrate that AtGPA1 directly interacts with PLDa1 and inhibits PLD catalytic activity.71 The functional implications of this inhibition have been delineated using PLDα1-deficient Arabidopsis.72 ABA promotes stomatal closing and this is mediated by PLD-generated phosphatidic acid which regulates the protein phosphatase ABI1.72 ABA also prevents stomatal opening in an AtGPA1-dependent manner.37 Wang and colleagues present genetic data consistent with a model in which AtGPA1 binds to PLD to mediate ABA-induced inhibition of stomatal opening.72 While the genetic data of Wang and colleagues are compelling, the biochemical data they provide are less straightforward. They demonstrate that PLDα1 binds specifically to the GDP-bound form of AtGPA1.71 Paradoxically, they also demonstrate that PLDα1 stimulates the steady state GTPase activity of AtGPA1.71 In light of our AtGPA1 biochemical data, this latter point is hard to interpret, given that accelerating steady state GTPase activity requires binding to the activated (GTP-bound) or transition-state form of AtGPA1.6 The effect of PLD was only minor: a 30% increase in GTPase activity at a 1:1 molar ratio.71 Further experiments should be done to clarify this supposed GAP activity of PLD. Wang and colleagues further provide an predicted molecular mechanism for AtGPA1/PLDα1 interaction by identifying a GPCR-like DRY motif in PLDα1 that, when mutated, abrogates this protein-protein interaction.71 The DRY motif in GPCRs is a crucial structural element that exists at the transmembrane helix III/intracellular loop interface involved in mediating contact between the receptor and G-protein.73 Wang and colleagues provide a sequence alignment purporting to show high sequence homology between PLDα1 and the second intracellular loop of chicken rhodopsin.71 However, it appears that they have misannotated these features and we present a corrected version in Figure 9. The PLDα1 ‘DR’ motif occurs within a predicted loop within the PLD catalytic domain (Fig. 9 and ref. 74). Thus, it is conceivable that a binding interaction at this site, essentially within the catalytic domain, would modulate PLD enzyme activity. However, classification of this as a bona fide “DRY motif” is misleading to the GPCR literature at-large, especially as the hydrophobic region downstream of these three amino acids in PLD is conserved through to bacterial phospholipases, whereas GPCRs are a eukaryotic-specific protein family.75 Hence, these should be considered evolutionarily- and functionally-distinct motifs.
Figure 9.
Alignment of Arabidopsis phospholipase Dα1 and rhodopsin based on primary sequence and predicted secondary structure elements. Primary sequences of Arabidopsis PLDα1 (GenBank accession: NP_188194), chicken rhodopsin (Swiss-Prot accession: P22328), bovine rhodopsin (Swiss-Prot accession: P02699), and the ostensible ‘Chicken Rhodopsin’ sequence (in italics) described by Zhao and Wang71 were aligned using ClustalW2 (www.ebi.ac.uk/Tools/clustalw2/index.html; ref. 85) and visualized using BoxShade3.21 (www.ch.embnet.org/software/BOX_form.html). The upper annotation denotes the predicted secondary structure of AtPLDα1 including α2 helix and β3 loop based on the crystal structure of the Samonella PLD ortholog Nuc,74 as well as inserts unique to Arabidopsis PLDα1. The lower annotation denotes the secondary structure of bovine rhodopsin based on the crystal structure.86 Note: the putative ‘DRY motif’ of Arabidopsis PLDα1 is predicted to be within a unique loop between the α2 helix and β3 loop of this enzyme. Note: the ‘chicken rhodopsin’ sequence described by Zhao and Wang71 is significantly different from the published sequence87; in fact, this rhodopsin sequence reported by Zhao and Wang appears to be the PLDα1 sequence but with an ERF tripeptide sequence affixed to the N-terminus. Secondary structure annotation was obtained from ‘author approved’ secondary structure annotations at the PDB (www.rcsb.org/pdb; PDB identifiers Salmonella typhimurium Nuc 1BYS and Bos taurus Rhodopsin PDB:1F88).
Conclusion
Unlike its mammalian Gα counterparts, the Arabidopsis thaliana AtGPA1 functions within the confines of a GTP hydrolysis-limited guanine nucleotide cycle. In mammalian systems, such as the photoresponse system within the retina, the GEF activity of agonist-activated GPCRs is critical for the onset of signal transduction (reviewed in ref. 76) and RGS proteins accelerate the termination of signaling, which often resets the system for further activation cycles.77 In particular within the retinal photoresponse system, cGMP-gated ion channels are constitutively open in the presence of high cGMP levels, until a photon activates the GPCR rhodopsin to activate Gα-transducin that stimulates cGMP phosphodiesterase and thus the destruction of cGMP. Thus, light-induced signaling turns off a constitutively-active signaling system. This is somewhat analogous to the plant system in which constitutively GTP-bound AtGPA1 is turned off in response to AtRGS1 activation. Sugar-mediated signaling in Arabidopsis, including hypocotyl development and growth arrest, appears to regulate AtGPA1 through an RGS-mediated GAP function of AtRGS1. The mammalian visual system is optimized to allow sub-second kinetic resolution; in contrast, the kinetics of Arabidopsis G-protein signal transduction are likely to be at the other end of the temporal scale. Plants are sessile and thus interact with and sense their environment in distinctly different ways than most organisms that possess G-proteins. Therefore the alternative functional and kinetic usage of G-protein signaling components by Arabidopsis may indicate a unique evolutionary adaptation to reflect the life cycle of plants.
Future studies will be needed to resolve in a definitive manner the molecular determinants of the AtGPA1 guanine nucleotide cycle and the basis for AtRGS1 regulation in response to binding of α-D-glucose. Similarly, a definitive biochemical analysis of AtGPA1 effectors and regulatory proteins needs to be conducted. These are great times to be studying G-protein signaling in such a unique environment.
Acknowledgements
Work in the Siderovski laboratory on Arabidopsis G-protein signaling is supported by National Institute of General Medical Sciences R01 GM082892 (to D.P.S.).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/7184
References
- 1.Temple BR, Jones AM. The plant heterotrimeric G-protein complex. Annu Rev Plant Biol. 2007;58:249–266. doi: 10.1146/annurev.arplant.58.032806.103827. [DOI] [PubMed] [Google Scholar]
- 2.Gao Y, Zeng Q, Guo J, Cheng J, Ellis BE, Chen JG. Genetic characterization reveals no role for the reported ABA receptor, GCR2, in ABA control of seed germination and early seedling development in Arabidopsis. Plant J. 2007;52:1001–1013. doi: 10.1111/j.1365-313X.2007.03291.x. [DOI] [PubMed] [Google Scholar]
- 3.Johnston CA, Temple BR, Chen JG, Gao Y, Moriyama EN, Jones AM, Siderovski DP, Willard FS. Comment on “A G protein coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid”. Science. 2007;318:914. doi: 10.1126/science.1143230. [DOI] [PubMed] [Google Scholar]
- 4.Moriyama EN, Strope PK, Opiyo SO, Chen Z, Jones AM. Mining the Arabidopsis thaliana genome for highly-divergent seven transmembrane receptors. Genome Biol. 2006;7:96. doi: 10.1186/gb-2006-7-10-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- 6.Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JG, Jones AM. AtRGS1 function in Arabidopsis thaliana. Methods Enzymol. 2004;389:338–350. doi: 10.1016/S0076-6879(04)89020-7. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Ji F, Xie H, Liang J, Zhang J. The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol. 2006;140:302–310. doi: 10.1104/pp.105.069872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma H, Yanofsky MF, Meyerowitz EM. Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc Natl Acad Sci USA. 1990;87:3821–3825. doi: 10.1073/pnas.87.10.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields TA, Casey PJ. Signalling functions and biochemical properties of pertussis toxin-resistant G-proteins. Biochem J. 1997;321:561–571. doi: 10.1042/bj3210561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson KM, Higashijima T, Smigel MD, Gilman AG. The influence of bound GDP on the kinetics of guanine nucleotide binding to G proteins. J Biol Chem. 1986;261:7393–7399. [PubMed] [Google Scholar]
- 12.Brandt DR, Ross EM. GTPase activity of the stimulatory GTP-binding regulatory protein of adenylate cyclase, Gs. Accumulation and turnover of enzyme-nucleotide intermediates. J Biol Chem. 1985;260:266–272. [PubMed] [Google Scholar]
- 13.Higashijima T, Ferguson KM, Sternweis PC, Smigel MD, Gilman AG. Effects of Mg2+ and the beta gamma-subunit complex on the interactions of guanine nucleotides with G proteins. J Biol Chem. 1987;262:762–766. [PubMed] [Google Scholar]
- 14.Linder ME, Pang IH, Duronio RJ, Gordon JI, Sternweis PC, Gilman AG. Lipid modifications of G protein subunits. Myristoylation of Go alpha increases its affinity for betagamma. J Biol Chem. 1991;266:4654–4659. [PubMed] [Google Scholar]
- 15.Seo HS, Jeong JY, Nahm MY, Kim SW, Lee SY, Bahk JD. The effect of pH and various cations on the GTP hydrolysis of rice heterotrimeric G-protein alpha subunit expressed in Escherichia coli. J Biochem Mol Biol. 2003;36:196–200. doi: 10.5483/bmbrep.2003.36.2.196. [DOI] [PubMed] [Google Scholar]
- 16.Seo HS, Kim HY, Jeong JY, Lee SY, Cho MJ, Bahk JD. Molecular cloning and characterization of RGA1 encoding a G protein alpha subunit from rice (Oryza sativa L. IR-36) Plant Mol Biol. 1995;27:1119–1131. doi: 10.1007/BF00020885. [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki Y, Kato T, Kaidoh T, Ishikawa A, Asahi T. Characterization of the putative alpha subunit of a heterotrimeric G protein in rice. Plant Mol Biol. 1997;34:563–572. doi: 10.1023/a:1005807010811. [DOI] [PubMed] [Google Scholar]
- 18.Ding L, Pandey S, Assmann SM. Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J. 2008;53:248–263. doi: 10.1111/j.1365-313X.2007.03335.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee YR, Assmann SM. Arabidopsis thaliana ‘extra-large GTP-binding protein’ (AtXLG1): a new class of G-protein. Plant Mol Biol. 1999;40:55–64. doi: 10.1023/a:1026483823176. [DOI] [PubMed] [Google Scholar]
- 20.Kimple RJ, Jones MB, Shutes A, Yerxa BR, Siderovski DP, Willard FS. Established and emerging fluorescence-based assays for G-protein function: heterotrimeric G-protein alpha subunits and regulator of G-protein signaling (RGS) proteins. Comb Chem High Throughput Screen. 2003;6:399–407. doi: 10.2174/138620703106298491. [DOI] [PubMed] [Google Scholar]
- 21.Yu JH. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J Microbiol. 2006;44:145–154. [PubMed] [Google Scholar]
- 22.Sun B, Firtel RA. A regulator of G protein signaling-containing kinase is important for chemotaxis and multicellular development in dictyostelium. Mol Biol Cell. 2003;14:1727–1743. doi: 10.1091/mbc.E02-08-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siderovski DP, Hessel A, Chung S, Mak TW, Tyers M. A new family of regulators of G-protein-coupled receptors? Curr Biol. 1996;6:211–212. doi: 10.1016/s0960-9822(02)00454-2. [DOI] [PubMed] [Google Scholar]
- 24.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2007;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 25.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 26.Marin EP, Krishna AG, Sakmar TP. Rapid activation of transducin by mutations distant from the nucleotide-binding site: evidence for a mechanistic model of receptor-catalyzed nucleotide exchange by G proteins. J Biol Chem. 2001;276:27400–27405. doi: 10.1074/jbc.C100198200. [DOI] [PubMed] [Google Scholar]
- 27.Onrust R, Herzmark P, Chi P, Garcia PD, Lichtarge O, Kingsley C, Bourne HR. Receptor and betagamma binding sites in the alpha subunit of the retinal G protein transducin. Science. 1997;275:381–384. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 28.Slessareva JE, Ma H, Depree KM, Flood LA, Bae H, Cabrera-Vera TM, Hamm HE, Graber SG. Closely related G-protein-coupled receptors use multiple and distinct domains on G-protein alpha-subunits for selective coupling. J Biol Chem. 2003;278:50530–50536. doi: 10.1074/jbc.M304417200. [DOI] [PubMed] [Google Scholar]
- 29.Bae H, Cabrera-Vera TM, Depree KM, Graber SG, Hamm HE. Two amino acids within the alpha4 helix of Galphai1 mediate coupling with 5-hydroxytryptamine1B receptors. J Biol Chem. 1999;274:14963–14971. doi: 10.1074/jbc.274.21.14963. [DOI] [PubMed] [Google Scholar]
- 30.Johnston CA, Siderovski DP. Structural basis for nucleotide exchange on Galphai subunits and receptor coupling specificity. Proc Natl Acad Sci USA. 2007;104:2001–2006. doi: 10.1073/pnas.0608599104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Iiri T, Herzmark P, Nakamoto JM, van Dop C, Bourne HR. Rapid GDP release from Gs alpha in patients with gain and loss of endocrine function. Nature. 1994;371:164–168. doi: 10.1038/371164a0. [DOI] [PubMed] [Google Scholar]
- 32.Posner BA, Mixon MB, Wall MA, Sprang SR, Gilman AG. The A326S mutant of Gialpha1 as an approximation of the receptor-bound state. J Biol Chem. 1998;273:21752–21758. doi: 10.1074/jbc.273.34.21752. [DOI] [PubMed] [Google Scholar]
- 33.Thomas TC, Schmidt CJ, Neer EJ. G-protein alpha o subunit: mutation of conserved cysteines identifies a subunit contact surface and alters GDP affinity. Proc Natl Acad Sci USA. 1993;90:10295–10298. doi: 10.1073/pnas.90.21.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS. G-protein signaling: back to the future. Cell Mol Life Sci. 2005;62:551–577. doi: 10.1007/s00018-004-4462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willard FS, Siderovski DP. Purification and in vitro functional analysis of the Arabidopsis thaliana regulator of G-protein signaling-1. Methods Enzymol. 2004;389:320–338. doi: 10.1016/S0076-6879(04)89019-0. [DOI] [PubMed] [Google Scholar]
- 36.Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science. 2001;292:2066–2069. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- 37.Wang XQ, Ullah H, Jones AM, Assmann SM. G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science. 2001;292:2070–2072. doi: 10.1126/science.1059046. [DOI] [PubMed] [Google Scholar]
- 38.Wang S, Narendra S, Fedoroff N. Heterotrimeric G protein signaling in the Arabidopsis unfolded protein response. Proc Natl Acad Sci USA. 2007;104:3817–3822. doi: 10.1073/pnas.0611735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandey S, Chen JG, Jones AM, Assmann SM. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 2006;141:243–256. doi: 10.1104/pp.106.079038. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 41.Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM. The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell. 2006;18:1226–1238. doi: 10.1105/tpc.105.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue Y, Batlle M, Hirsch JP. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. Embo J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell. 2004;16:293–299. doi: 10.1016/j.molcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 45.Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 46.Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 47.Palmer RK. The pharmacology and signaling of bitter, sweet and umami taste sensing. Mol Interv. 2007;7:87–98. doi: 10.1124/mi.7.2.9. [DOI] [PubMed] [Google Scholar]
- 48.Jang HJ, Kokrashvili Z, Theodorakis MJ, Carlson OD, Kim BJ, Zhou J, Kim HH, Xu X, Chan SL, Juhaszova M, Bernier M, Mosinger B, Margolskee RF, Egan JM. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc Natl Acad Sci USA. 2007;104:15069–15074. doi: 10.1073/pnas.0706890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margolskee RF, Dyer J, Kokrashvili Z, Salmon KS, Ilegems E, Daly K, Maillet EL, Ninomiya Y, Mosinger B, Shirazi-Beechey SP. T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc Natl Acad Sci USA. 2007;104:15075–15080. doi: 10.1073/pnas.0706678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon MI, Strathmann MP, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 51.Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malbon CC. G proteins in development. Nat Rev Mol Cell Biol. 2005;6:689–701. doi: 10.1038/nrm1716. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Yue Y, Li B, Nie Y, Li W, Wu WH, Ma L. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- 54.Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 55.Liu X, Yue Y, Li W, Ma L. Response to Comment on “A G Protein Coupled Receptor Is a Plasma Membrane Receptor for the Plant Hormone Abscisic Acid”. Science. 2007;318:914. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- 56.Illingworth CJ, Parkes KE, Snell CR, Mullineaux PM, Reynolds CA. Criteria for confirming sequence periodicity identified by Fourier transform analysis: Application to GCR2, a candidate plant GPCR? Biophys Chem. 2008;133:28–35. doi: 10.1016/j.bpc.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 58.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 59.Pandey S, Assmann SM. The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell. 2004;16:1616–1632. doi: 10.1105/tpc.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 2004;135:907–915. doi: 10.1104/pp.104.038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bayle D, Weeks D, Sachs G. Identification of membrane insertion sequences of the rabbit gastric cholecystokinin-A receptor by in vitro translation. J Biol Chem. 1997;272:19697–19707. doi: 10.1074/jbc.272.32.19697. [DOI] [PubMed] [Google Scholar]
- 62.Robelek R, Lemker ES, Wiltschi B, Kirste V, Naumann R, Oesterhelt D, Sinner EK. Incorporation of in vitro synthesized GPCR into a tethered artificial lipid membrane system. Angew Chem Int Ed Engl. 2007;46:605–608. doi: 10.1002/anie.200602231. [DOI] [PubMed] [Google Scholar]
- 63.Dascal N. Ion-channel regulation by G proteins. Trends Endocrinol Metab. 2001;12:391–398. doi: 10.1016/s1043-2760(01)00475-1. [DOI] [PubMed] [Google Scholar]
- 64.Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Assmann SM. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature. 2003;423:651–654. doi: 10.1038/nature01643. [DOI] [PubMed] [Google Scholar]
- 65.Lein W, Saalbach G. Cloning and direct G-protein regulation of phospholipase D from tobacco. Biochim Biophys Acta. 2001;1530:172–183. doi: 10.1016/s1388-1981(00)00182-7. [DOI] [PubMed] [Google Scholar]
- 66.Munnik T, Arisz SA, De Vrije T, Musgrave A. G Protein Activation Stimulates Phospholipase D Signaling in Plants. Plant Cell. 1995;7:2197–2210. doi: 10.1105/tpc.7.12.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.den Hartog M, Musgrave A, Munnik T. Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: a role for phospholipase C and D in root hair deformation. Plant J. 2001;25:55–65. doi: 10.1046/j.1365-313x.2001.00931.x. [DOI] [PubMed] [Google Scholar]
- 68.Miles GP, Samuel MA, Jones AM, Ellis BE. Mastoparan rapidly activates plant MAP kinase signaling independent of heterotrimeric G proteins. Plant Physiol. 2004;134:1332–1336. doi: 10.1104/pp.103.037275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones S, Howl J. Biological applications of the receptor mimetic peptide mastoparan. Curr Protein Pept Sci. 2006;7:501–508. doi: 10.2174/138920306779025585. [DOI] [PubMed] [Google Scholar]
- 70.Avigan J, Murtagh JJ, Jr, Stevens LA, Angus CW, Moss J, Vaughan M. Pertussis toxin-catalyzed ADP-ribosylation of G(o) alpha with mutations at the carboxyl terminus. Biochemistry. 1992;31:7736–7740. doi: 10.1021/bi00148a039. [DOI] [PubMed] [Google Scholar]
- 71.Zhao J, Wang X. Arabidopsis phospholipase Dalpha1 interacts with the heterotrimeric G-protein alpha-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Chem. 2004;279:1794–1800. doi: 10.1074/jbc.M309529200. [DOI] [PubMed] [Google Scholar]
- 72.Mishra G, Zhang W, Deng F, Zhao J, Wang X. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science. 2006;312:264–266. doi: 10.1126/science.1123769. [DOI] [PubMed] [Google Scholar]
- 73.Rovati GE, Capra V, Neubig RR. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol Pharmacol. 2007;71:959–964. doi: 10.1124/mol.106.029470. [DOI] [PubMed] [Google Scholar]
- 74.Stuckey JA, Dixon JE. Crystal structure of a phospholipase D family member. Nat Struct Biol. 1999;6:278–284. doi: 10.1038/6716. [DOI] [PubMed] [Google Scholar]
- 75.Rompler H, Staubert C, Thor D, Schulz A, Hofreiter M, Schoneberg T. G protein-coupled time travel: evolutionary aspects of GPCR research. Mol Interv. 2007;7:17–25. doi: 10.1124/mi.7.1.5. [DOI] [PubMed] [Google Scholar]
- 76.Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 77.Zhong H, Wade SM, Woolf PJ, Linderman JJ, Traynor JR, Neubig RR. A spatial focusing model for G protein signals. Regulator of G protein signaling (RGS) protien-mediated kinetic scaffolding. J Biol Chem. 2003;278:7278–7284. doi: 10.1074/jbc.M208819200. [DOI] [PubMed] [Google Scholar]
- 78.Afshar K, Willard FS, Colombo K, Johnston CA, McCudden CR, Siderovski DP, Gonczy P. RIC-8 is required for GPR-1/2-dependent Galpha function during asymmetric division of C. elegans embryos. Cell. 2004;119:219–230. doi: 10.1016/j.cell.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 79.Willard FS, Kimple AJ, Johnston CA, Siderovski DP. A direct fluorescence-based assay for RGS domain GTPase accelerating activity. Anal Biochem. 2005;340:341–351. doi: 10.1016/j.ab.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 80.Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 81.Insall R. The Dictyostelium genome: the private life of a social model revealed? Genome Biol. 2005;6:222. doi: 10.1186/gb-2005-6-6-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnston CA, Siderovski DP. Receptor-mediated activation of heterotrimeric G-proteins: current structural insights. Mol Pharmacol. 2007;72:219–230. doi: 10.1124/mol.107.034348. [DOI] [PubMed] [Google Scholar]
- 83.Coleman DE, Sprang SR. Crystal structures of the G protein Gialpha1 complexed with GDP and Mg2+: a crystallographic titration experiment. Biochemistry. 1998;37:14376–14385. doi: 10.1021/bi9810306. [DOI] [PubMed] [Google Scholar]
- 84.Rolland F, Winderickx J, Thevelein JM. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002;2:183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 85.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 87.Takao M, Yasui A, Tokunaga F. Isolation and sequence determination of the chicken rhodopsin gene. Vision Res. 1988;28:471–480. doi: 10.1016/0042-6989(88)90169-1. [DOI] [PubMed] [Google Scholar]