Abstract
The cross-talk between plant disease resistance and development is fundamental to understanding systemic physiological processes during pathogen attack. Our previous study showed that the Arabidopsis GH3.5 gene acts as a bifunctional modulator of the salicylic acid (SA)-mediated resistance and the auxin-mediated susceptibility during the Arabidopsis-Pseudomonas syringae interaction as well as development. Here, we further study the role and mechanism of GH3.5 involved in the SA-dependent defense pathway. Transcript and histochemical analysis of the GH3.5 promoter::GUS reporter expression indicate that GH3.5 is expressed with a strong temporal and spatial manner with predominant expression in the divisional tissues. Upon bacterial challenge, GUS activity is induced in the junction tissue around the infiltrated zone with higher levels in the vasculature with a pattern different between the incompatible and compatible interactions. Exogenous SA application enhances disease resistance in the activation-tagged mutant gh3.5-1D, while the GH3.5-mediated defense enhancement is depleted in the SA deficient gh3.5-1D/NahG double mutant, indicating that GH3.5 modulates defense response through the SA-dependent pathway. Furthermore, bacterial growth in the gh3.5-1D/npr1 double mutant treated with SA indicates that GH3.5 enhances the SA-mediated defense response through both NPR1-dependent and independent pathways.
Key words: GH3.5, salicylic acid, defense response, double mutants, NPR1, NahG, Pseudomonas syringae
Introduction
It has been widely recognized that SA plays a central role in plant defense against pathogens and is required for the induction of a set of PR genes and synthesis of defense compounds.1 Exogenous SA application can induce a set of PR genes and establish the systematic acquired resistance (SAR), resulting in broad-spectrum disease resistance.2 Transgenic plants expressing the NahG gene, encoding a salicylate hydroxylase that catalyzes the conversion of SA to catechol, fail to accumulate SA after pathogen infection and are compromised in SAR, basal resistance and some R gene-mediated resistance.3,4 Furthermore, the Arabidopsis sid1/eds5 and sid2/eds16 mutants deficient in pathogen-induced SA accumulation are also impaired in SAR, certain R gene-mediated resistance and basal resistance.5,6 Extensive studies have also shown that the NPR1 gene (also known as NIM1 and SAI1) functions as the key regulator of the SA-mediated SAR. The mutants of the gene, npr1/nim1/sai1, lost the expression of SA-induced PR genes and SAR.7–10
The GH3 gene family, an early auxin-responsive gene group, was first isolated by differential screening from etiolated seedling hypocotyls of soybean (Glycine Max) after treatment with 2,4-dichlorophenoxyacetic acid (2,4-D).11,12 One of Group II GH3 proteins of Arabidopsis, GH3.5 (At4g27260), adenylated both indole acetic acid (IAA) and salicylic acid (SA) in vitro,13,14 suggesting that GH3.5 could potentially function in modulating and integrating both the auxin and SA signaling pathways. We conducted extensive genetic, molecular and biochemistry analysis, and demonstrated that GH3.5 acts as a bifunctional modulator of SA and auxin signaling during the Arabidopsis-Pseudomonas syringae (P. syringae) interaction.15 The activation-tagged mutant gh3.5-1D that overexpresses GH3.5 accumulated high SA levels and increased expression of PR-1 in local and systemic tissues in response to avirulent P. syringae. By contrast, two T-DNA insertion mutants of GH3.5 partially compromises the systemic acquired resistance associated with diminished PR-1 expression in systemic tissues. The gh3.5-1D mutant also accumulated high levels of free IAA after pathogen infection and impairs different R genes-mediated resistance, revealing another dimension to the complex and dynamic plant-pathogen interaction.15 A similar activation-tagged mutant of the GH3.5 gene, wes1-D, was also recently reported to accumulate SA and exhibit enhanced disease resistance to Pst DC3000 at the flowering stage with spray-inoculation under the long day conditions, which also exhibited the altered light response.16,17
Recently, GH3.12/PBS3/GDG1/WIN3, a group III GH3 member, was shown to positively regulate SA-dependent disease resistance.18–20 Taken together, these studies indicate that some members of the GH3 family play a critical role in SA signaling and induced defense responses. However, how these GH3s regulate the SA-mediated defense response is still largely unknown. For example, we have shown that although SA accumulation and the expression of PR-1 were elevated in local and systemic tissues in response to avirulent pathogens, the R gene-mediated local resistance was compromised in gh3.5-1D due to counteracting by the auxin-mediated susceptibility,15 as the elevated auxin levels within host tissue promote P. syringae virulence and the type III effector AvrRpt2 may be one of the virulence factors of P. syringae that modulate host auxin physiology to promote disease.21 It is also intriguing that the rice GH3.8 gene plays a role in the SA- and jasmonate-independent basal immunity through suppressing expansin expression, indicating that the cell wall is actively involved in the plant immunity.22
In this work, we investigate the roles of GH3.5 in SA-dependent defense pathway with double mutants (transgenes) gh3.5-1D/Nahg and gh3.5-1D/npr1, and demonstrate that GH3.5 modulates defense response through SA-dependent and both NPR1-dependent and independent pathways.
Results and Discussion
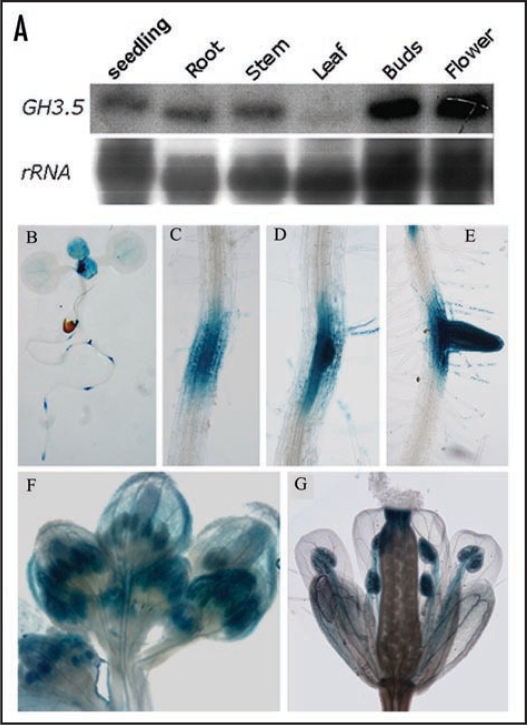
To assess developmental regulation of the GH3.5 gene, we detected the GH3.5 transcript in young seedlings and different organs of the adult wild-type plant by northern blot analysis. As shown in Figure 1A, except low level in the rosette leaves, high expression levels of GH3.5 were detected in one-week-old whole seedling, root, stem, buds and blooming flowers of the adult plant. This pattern is slightly different from the previous observation with RT-PCR Southern blot which showed that GH3.5 was expressed at a relatively high level in rosette leaves.16 In order to further confirm the GH3.5 expression pattern, we developed transgenic plants (GH3.5-GUS) carrying a GH3.5 promoter-GUS reporter fusion gene, and detected GUS activity in different tissues. In one-week-old seedlings, GUS activity was mainly centralized at the meristems, newly-born true leaves, root tips, lateral root primordia and developing lateral roots (Fig. 1B–E). GUS activity was also detected at the vascular bundles of cotyledons. In the buds, GUS was mainly stained at sepals and developing stamens (Fig. 1F). In the blooming flowers, GUS activity was mostly distributed at stigma, filaments and anthers (Fig. 1G). Taken together, these results demonstrate that GH3.5 is expressed with a strong temporal and spatial manner with predominant expression in the divisional tissues where auxin is produced and functions.
Figure 1.
Expression pattern of GH3.5. (A) Northern blot analysis of GH3.5 in different tissues of Col-0. (B) GUS activity in 7-day-old seedling of the GH3.5-GUS transgenic plant. (C–E) GUS activity during lateral root development in the GH3.5-GUS plant. (F) GUS activity in buds of the GH3.5-GUS plant. (G) GUS activity in the blooming flower of the GH3.5-GUS plant.
To examine roles of GH3.5 in the SA-dependent defense pathway, we first examined the exogenous SA-induced disease resistance (SAR) in gh3.5-1D. Wildtype and gh3.5-1D plants were first treated with 1 mM SA or buffer (mock) and inoculated with virulent Pst DC3000 two days later. We observed that SA treatment alleviated disease symptom in gh3.5-1D plants compared with wildtype plants (Fig. 2A). Bacterial growth titer decreased 0.73 Log in the SA-treated Col-0 plants compared to the mock-treated plants at day 3 post inoculation (dpi), confirming that SA could trigger a disease resistance response in wildtype plants as expected (Fig. 2B). Interestingly, the SA-induced resistance was indeed stronger (p < 0.05) in gh3.5-1D compared with the wildtype, which exhibied a 1.5 Log decrease of bacterial growth titer compared to the mock-treated plants at 3 dpi. The result demonstrates that GH3.5 positively modulates SA signaling, leading to a slight but reliable increased resistance to virulent pathogen in gh3.5-1D after exogenous SA treatment. Therefore, this result further supported the previous observation that the pathogen-induced SAR was indeed stronger induced in gh3.5-1D than the wildtype but was counteracted by the simultaneously augmented auxin-mediated susceptibility, leading to the normal resistance outcome in term of bacterial growth.15
Figure 2.
SA-induced disease resistance in Col-0 and gh3.5-1D. (A) Disease symptom of Pst DC3000 in mock and SA-treated Col-0 (left) and gh3.5-1D (+/−) (right). Five-week-old plants of the wildtype and mutants were sprayed with 1 mM SA and further inoculated with Pst DC3000 at a dose of 105 cfu ml−1 (OD600 = 0.0002). All controls were infiltrated with 10 mM MgCl2 for mock inoculation. Photos were taken at 3 dpi. Similar results were observed in two independent experiments. (B) Growth of Pst DC3000 in SA-treated leaves of Col-0 and gh3.5-1D. Bacterial growth assay was performed at 0 and 3 days after inoculation in Col-0 and gh3.5-1D (+/−). All values are means ± SE (n = 6). The SA-induced resistance was significantly stronger (p < 0.05) in gh3.5-1D than the wldtype. cfu, colony-forming units. Similar results were observed in two independent experiments.
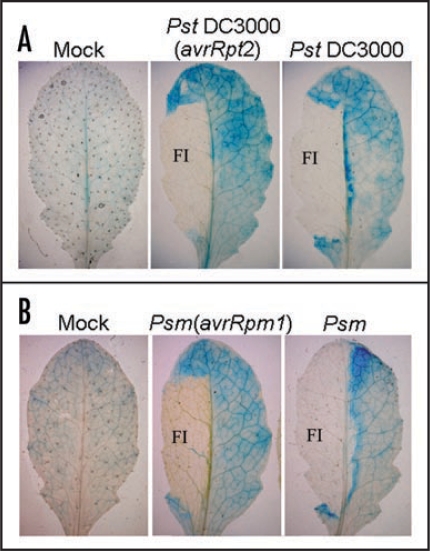
GH3.5 is inducible by P. syringae with different induction patterns by avirulent and virulent strains in wildtype plants.15 To further examine the induction pattern of GH3.5, GUS activity was examined histochemically in the GH3.5-GUS transgenic plants. We observed that GH3.5-GUS was expressed at a low basal level in MgCl2-treated leaves (Fig. 3A and B). When challenged with bacteria, GUS activity was induced in the junction tissue around the infiltrated zone with higher levels in the vasculature in response to both virulent and avirulent pathogens. Interestingly, GUS activity was mainly centralized around the chlorotic areas, with limited extension into the vasculature in the compatible interaction. Taken together, the different induction patterns of GH3.5 by the virulent and avirulent pathogens further support the hypothesis that GH3.5 plays different roles in the compatible and incompatible interactions.15
Figure 3.
Histochemical assay of the expression pattern of the GH3.5-GUS reporter in responses to pathogen. (A) Induction of the GH3.5-GUS fusion reporter by Pst DC3000(avrRpt2) and Pst DC3000 at 107 cfu ml−1 at 3 dpi. FI, filter-infected areas. (B) Induction of the GH3.5-GUS fusion reporter by Psm(avrRpm1) and Psm at 107 cfu ml−1 at 3 dpi. Leaves were infiltrated with 10 mM MgCl2 for mock inoculation (A and B).
Since the activation of the SA pathway is associated with the expression of certain PR genes such as PR-1,2 we further analyzed the expression of PR-1 induced by exogenous SA in both gh3.5-1D and wildtype plants. As shown in Figure 4A, the kinetics of PR-1 induction was altered in gh3.5-1D compared with the wildtype after SA treatment. In contrast to a prolonged elevation of the PR-1 transcript from 6 to 48 h in the wildtype control, the SA-mediated induction of the PR-1 gene exhibited a sharper pattern with a peak at 12 h and no PR-1 expression at 48 h in both heterozygous and homozygous gh3.5-1D plants. These results indicated that GH3.5 regulates PR-1 in response to SA treatment, with a significant effect on the kinetics of PR-1 expression. This kind of defense gene induction was previously observed during incompatible interactions in bean and parsley cells,25,26 suggesting that this kind of expression pattern may be related to the establishment of an efficient resistance.
Figure 4.
GH3.5 modulates SA-dependent defense response. (A) Northern blot analysis of PR-1 induction by exogenous application of SA (0.5 mM) over a time course of 0 to 48 h after treatment in gh3.5-1D heterozygous (gh3.5-1D+/−) and homozygous (gh3.5-1D−/−) plants, in comparison with Col-0 plants. The experiment was biologically repeated once. (B) Expression of PR-1 in NahG, gh3.5-1D/NahG double mutant and gh3.5-1D plants after infection with Psm(avrRpm1) at 107 cfu ml−1. Leaves were collected at 0 and 48 hpi. The experiments were repeated once with similar results. (C) Growth of Pst DC3000 in Col-0, NahG and gh3.5-1D/NahG plants after pre-treatment with SA (1 mM) or buffer (mock). Bacterial titers were repeated twice with similar results. All values are the mean ± SE (n = 6).
To determine whether the GH3.5-enhanced defense depends on SA accumulation, we generated a double mutant between the gh3.5-1D mutant and the NahG transgenic plants known to be defective in SA accumulation.3,4 The expression of PR-1 was first analyzed in these plants after inoculation with Psm(avrRpm1). As shown in Figure 4B, PR-1 was only slightly induced in both NahG and gh3.5-1D/NahG plants, which is in sharp contrast to a strong PR-1 induction in gh3.5-1D, demonstrating that the increased expression of PR-1 in gh3.5-1D is dependent on SA accumulation. In addition to the reduced PR-1 induction, bacterial growth assays revealed that both the SA-induced SAR and basal resistance were abolished in gh3.5-1D/NahG plants like in NahG plants at 3 dpi (Fig. 4C; reviewed in ref. 15). These results confirmed that the enhancement of defense responses by overexpressing GH3.5 indeed depends on SA accumulation.
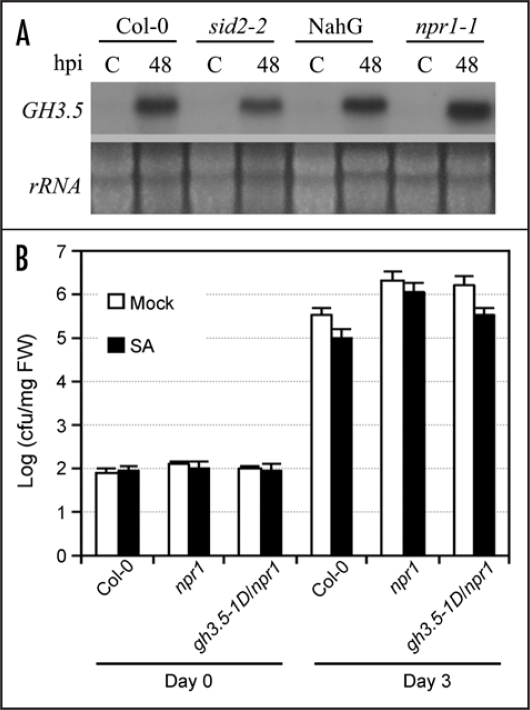
We have shown that GH3.5 is inducible by pathogen and SA.15 However, the induction of GH3.5 by Psm(avrRpm1) in NahG and sid2-227 plants, both accumulating low levels of SA in response to pathogen, was quite same to that in the equivalent wildtype plants (Fig. 5A). Similar induction of GH3.5 was also observed in the npr1 mutant. These results further support the postulation that the pathogen-induced expression of the GH3.5 gene is mediated most likely by an SA-independent pathway, and plays an important role in strengthening the SA pathway.15 To further address the GH3.5-mediated SA pathway enhancement in SAR, we developed another double mutant gh3.5-1D/npr1 since NPR1 is a major regulator of SAR,28 and conducted the SA-mediated SAR assay. As shown in Figure 5B, there was a 0.51 Log (p < 0.05) decrease of bacterial growth in gh3.5-1D/npr1 plants than in npr1 plants after SA treatment, a growth value higher than that in gh3.5-1D treated with SA (Fig. 2), suggesting that SA treatment induced resistance through both the NPR1-dependent and independent pathways in gh3.5-1D.
Figure 5.
SA-independent induction of GH3.5 and NPR1-dependent and independent SA-induced defense. (A) The induction of GH3.5 by Psm(avrRpm1) at 107 cfu ml−1 in Col-0, NahG, sid2-2 and npr1-1 plants, indicating that the GH3.5 induction was not affected by SA deficiency in these mutants. (B) Growth of Pst DC3000 in Col-0, npr1 and gh3.5-1D/npr1 plants after pre-treatment with SA (1 mM) or buffer. Bacterial titers were repeated twice with similar results. All values are the mean ± SE (n = 6).
Although extensively studies have been done on SA signaling, little is known about how SA acts. For instance, it is well established that the NPR1-regulated expression of PR genes is required for the induction of SAR,9,28 but understanding of the molecular mechanism underlying SAR against a broad spectrum of pathogens is limited. The recent finding that methyl salicylate is a mobile signal for SAR opens a door to these questions.29 Our data has revealed that GH3.5 plays an important role in the regulation of SA signaling in plant defense, since gh3.5-1D exhibited a stronger SA-induced disease resistance accompanied by a greatly altered kinetics of PR-1 expression. Furthermore, the PR-1 induction was no longer strengthened in the gh3.5-1D/NahG double mutant that exhibited no SAR. These results indicate that GH3.5 enhances the SA-mediated disease resistance, providing the further evidence that GH3.5 act as a positive regulator of the SA pathway in response to pathogen. Interestingly, the GH3.5-enhanced SA pathway is both NPR1-dependent and independent since the gh3.5-1D/npr1 double mutant still exhibited partial SAR. Consistent with this, our previous microarray analysis revealed some NPR1-dependnet WRKY and TGA transcription factors, and NPR1-independent α-DOX1.15 Our current study also adds evidence that the SA-dependent defense responses are either NPR1-depdendent or independent.30–34 How these signaling pathways are integrated to defense pathogens is still a big challenge to plant biologists.
Materials and Methods
Plant materials and growth conditions.
The activation-tagged mutant gh3.5-1D was previously described.15 The sid2 mutant was provided by Prof. Fred Ausubel, the npr1 mutant was provided by Prof. Xinnian Dong. Seeds were surface sterilized and germinated on 1/2 MS agar medium. Plants were grown in a growth room under 22–23°C, 60% relative humidity, 85 µmol s−1m−2 fluorescent illumination, with 9/15 h day/night for pathogen inoculation, and 16/8 h day/night for physiological analysis.
Construction of double mutant.
For construction of the gh3.5-1D/NahG double mutant, gh3.5-1D was crossed with NahG (obtained from Novartis), a transgenic line expressing a bacterial salicylate hydroxylase that is unable to accumulate SA.4 The double mutant plants with gh3.5-1D and NahG were selected in F3 progenies. Homozygous NahG plants were confirmed by PCR. These double mutant plants are morphologically similar to gh3.5-1D. For construction of the double mutant gh3.5-1D/npr1, the gh3.5-1D was crossed with the npr1-1 mutant.9 The gh3.5-1D/npr1 plants were selected in F3 progenies and the mutation of npr1 was confirmed with cleaved-amplified polymorphic sequence marker using restriction endonuclease NlaIII. The gh3.5-1D/npr1 double mutant plants are morphologically similar to gh3.5-1D.
Promoter activity.
For the GH3.5 promoter-GUS fusion reporter, the PCR primers 5′-TTAGTAAGTTTCAGTCGACGTTCTAG A-3′ and 5′-TTGGATCCTCAGGCGTGGTTTAAGAG-3′, were used to amplify the 1.6-kb upstream region of the GH3.5 from genomic DNA. The PCR product was inserted into the pBI101 vector. The construct was then introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis (Col-0) by floral dip to produce more than 50 independent GH3.5-GUS transgenic plants. Homozygous transgenic plants were selected in the progenies. Histochemical assay for GUS activity in GH3.5-GUS plants was performed as described.23
Bacterial strains, inoculation and disease assessment.
Pseudomonas syringae strains and inoculation were previously described.15 In brief, bacterial pathogens grown in King's B (KB) medium were collected and re-suspended in 10 mM MgCl2. Leaves of 5-week-old plants were infiltrated with a bacterial suspension. All procedural controls were infiltrated with 10 mM MgCl2 for mock inoculation. Bacterial growth assay was performed as described.24 Except where otherwise noted, heterozygous gh3.5-1D(+/−) plants were used for all of bacterial growth experiments.
SA treatment.
Salicylic acid (Sigma-Aldrich, MO) was dissolve in distilled water. Five-week-old plants of the wild-type and mutants were sprayed with 1 mM SA containing 0.01% Silwett L-77 or buffer alone. Two days later, plants in each treatment were further inoculated with Pst DC3000 at a dose of 105 cfu ml−1 (OD600 = 0.0002). Bacterial growth titers were counted at 0 and 3 days after inoculation.
Northern blot analysis.
Total RNA was isolated from leaf tissues using TRIzol reagent according to the manufacturer's protocol (GIBCO BRL). Each 10 µg RNA of samples were separated on a formaldehyde-agarose gel, then blotted to Hybond-N+ membranes (Amersham). A 353-bp fragment of the GH3.5 3′ non-coding region was labeled with [α-32P]dCTP using a random primer labeling kit (Takara) for hybridization and autoradiograph as previously described.15 Northern blot was also performed to determine the transcript levels of the pathogenesis-related gene PR-1. The filters were reprobed with a 2.5-kb fragment of Arabidopsis 18S rRNA for loading normalization.
Acknowledgements
We thank Xinnan Dong for the npr1 mutant, Fred Ausubel for the sid2 mutant. This work was supported by the National Natural Science Foundation of China (30730064 and 30721061), and the Chinese Academy of Sciences (KSCX2-YW-N-007).
Abbreviations
- 2,4-D
2,4-dichlorophenoxyacetic acid
- IAA
indole acetic acid
- KB
king's B
- PR
pathogenesis-related
- R
resistance
- SA
salicylic acid
- SAR
systematic acquired resistance
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5748
References
- 1.Dempsey DA, Shah J, Klessig DF. Salicylic acid and disease resistance in plants. Crit Rev Plant Sci. 1999;18:547–575. [Google Scholar]
- 2.Uknes S, Mauch Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 4.Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negmtto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1125–1247. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 5.Rogers EE, Ausubel FM. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell. 1997;9:305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nawrath C, Métraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 10.Shah J, Tsui F, Klessig DF. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant-Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- 11.Hagen G, Guilfoyle TJ. Rapid induction of selective transcription by auxin. Mol Cell Biol. 1985;5:1197–1203. doi: 10.1128/mcb.5.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagen G, Kleinschmidt AJ, Guilfoyle TJ. Auxin regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta. 1984;162:147–153. doi: 10.1007/BF00410211. [DOI] [PubMed] [Google Scholar]
- 13.Staswick PE, Tiryaki I, Rowe M. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007;145:450–464. doi: 10.1104/pp.107.106021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem. 2007;282:10036–10046. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]
- 17.Park JE, Seo PJ, Lee AK, Jung JH, Kim YS, Park CM. An Arabidopsis GH3 Gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol. 2007;48:1236–1241. doi: 10.1093/pcp/pcm086. [DOI] [PubMed] [Google Scholar]
- 18.Jagadeeswaran G, Raina S, Acharyal BR, Maqbool SB, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R. Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 2007;51:234–246. doi: 10.1111/j.1365-313X.2007.03130.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee MW, Lu H, Jung HW, Greenberg JT. A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. Mol Plant Microbe Interact. 2007;20:1192–2120. doi: 10.1094/MPMI-20-10-1192. [DOI] [PubMed] [Google Scholar]
- 20.Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L, Wildermuth MC, Innes RW. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007;144:1144–1156. doi: 10.1104/pp.107.097691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA. 2007;104:20131–20136. doi: 10.1073/pnas.0704901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independentbasal immunity in rice. Plant Cell. 2008 doi: 10.1105/tpc.107.055657. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katagiri F, Thilmony R, He SY. The Arabidopsis thaliana-Pseudomonas syringae interaction. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedrick SA, Bell JN, Boller T, Lamb CJ. Chitinase cDNA 49 cloning and mRNA induction by fungal elicitor, wounding, and infection. Plant Physiol. 1988;86:182–186. doi: 10.1104/pp.86.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somssich IE, Bollmann J, Hahlbrock K, Kombrink E, Schultz W. Differential early activation of defense-related genes in elicitor-treated parsley cells. Plant Mol Biol. 1989;12:227–234. doi: 10.1007/BF00020507. [DOI] [PubMed] [Google Scholar]
- 27.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 28.Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science. 2007;318:113–116. doi: 10.1126/science.1147113. [DOI] [PubMed] [Google Scholar]
- 30.Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol. 2003;51:21–37. doi: 10.1023/a:1020780022549. [DOI] [PubMed] [Google Scholar]
- 31.Desveaux D, Subramaniam R, Després C, Mess JN, Lévesque C, Fobert PR, Dangl JL, Brisson N. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell. 2004;6:229–240. doi: 10.1016/s1534-5807(04)00028-0. [DOI] [PubMed] [Google Scholar]
- 32.Uquillas C, Letelier I, Blanco F, Jordana X, Holuigue L. NPR1-independent activation of immediate early salicylic acid-responsive genes in Arabidopsis. Mol Plant Microbe Interact. 2004;17:34–42. doi: 10.1094/MPMI.2004.17.1.34. [DOI] [PubMed] [Google Scholar]
- 33.Blanco F, Garreto V, Frey N, Dominguez C, Perez-Acle T, Van der Straeten D, Jordana X, Holuigue L. Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol Biol. 2005;59:927–944. doi: 10.1007/s11103-005-2227-x. [DOI] [PubMed] [Google Scholar]
- 34.Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogen. 2006;2:123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]