Abstract
The plant signaling hormones salicylic acid (SA) and jasmonic acid (JA) are regulators of inducible defenses that are activated upon pathogen or insect attack. Cross-talk between SA- and JA-dependent signaling pathways allows a plant to finely tune its response to the attacker encountered. In Arabidopsis, pharmacological experiments revealed that SA exerts a strong antagonistic effect on JA-responsive genes, such as PDF1.2, indicating that the SA pathway can be prioritized over the JA pathway. SA-mediated suppression of the JA-responsive PDF1.2 promoter was exploited for setting up a genetic screen aiming at the isolation of signal transduction mutants that are impaired in this cross-talk mechanism. The PDF1.2 promoter was fused to the herbicide resistance gene BAR to allow for life/death screening of a population of mutagenized transgenic plants. Non-mutant plants should survive herbicide treatment when methyl jasmonate (MeJA) is applied, but suppression of the JA response by SA should be lethal in combination with the herbicide. Conversely, crucial SA/JA cross-talk mutants should survive the combination treatment. SA effectively suppressed the expression of the PDF1.2::BAR transgene. However, suppression of the BAR gene did not result in suppression of herbicide resistance. Hence, a screening method based on quantitative differences in the expression of a reporter gene may be better suited to identify SA/JA cross-talk mutants. Here, we demonstrate that the PDF1.2::GUS reporter will be excellently suited in this respect.
Key words: plant defense, salicylic acid, jasmonic acid, cross-talk, mutant screen, Arabidopsis
Introduction
Arabidopsis thaliana has been adopted as a model organism for biologists worldwide, rendering a wealth of well-characterized mutants. The Arabidopsis genome was sequenced in 2000, and at present projects are underway to determine the function of all ∼25.500 genes by 2010.1 How these genes are regulated is a further question to be addressed. It has become increasingly clear that gene regulation through signal transduction pathways is subject to regulatory networks, rather than linear pathways. Therefore, multiple factors can contribute to the final outcome of a gene-regulated process. The major focus of future Arabidopsis research will be to understand the dynamic properties of the genes and networks that control plant functioning. For the dissection of such complex pathways, the identification of mutants in regulatory components remains essential.
Plants have evolved sophisticated defense mechanisms to resist invasion by deleterious organisms. The plant hormones salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) emerged as key players in the regulation of the defense signaling networks involved.2–6 Their pathways regulate defense responses that are differentially effective against specific types of attackers. Pathogens with a biotrophic lifestyle are generally more sensitive to SA-dependent responses, whereas necrotrophic pathogens and herbivorous insects are commonly deterred by JA/ET-dependent defenses.7–10
There is ample evidence that SA and JA signaling pathways cross-communicate, providing the plant with a highly flexible defense signaling network.11–13 Pathway cross-talk is thought to optimize the defense reaction to a particular attacker by enhancing the appropriate response, while suppressing suboptimal reactions. Indeed, trade-offs between defense signaling pathways have been demonstrated in several plant species.12 For instance, Spoel et al.,14 demonstrated that simultaneous infection of Arabidopsis with a biotrophic and a necrotrophic pathogen resulted in impaired resistance to the necrotrophic pathogen, demonstrating that the SA pathway that was activated by the biotroph suppressed the level of JA-dependent resistance against the necrotroph. Over the past years, several molecular players in SA/JA cross-talk have been identified.13 The regulatory protein NPR1 was found to be essential for both activation of SA-responsive PR gene expression and SA-mediated suppression of JA-responsive genes.15,16 In addition, the WRKY70 transcription factor and glutaredoxin GRX480 both have been shown to play a role in the regulation of SA/JA cross-talk.17,18 However, the interactions between these signaling components in the regulation of SA/JA cross-talk remain largely undetermined.
The use of forward genetic screens has been successful in identifying and characterizing many genes that have essential roles in the regulation of plant defense responses.19–21 So far, no mutant screens have been performed with the specific aim to unravel the molecular mechanism underlying SA/JA cross-talk. Hence, an approach specifically targeting SA/JA cross-talk mutants will be highly instrumental in the identification of signaling components that act in the antagonism on JA signaling in Arabidopsis. Here, we describe attempts to develop a system that allows efficient screening of mutants in which the suppression of JA-induced gene expression by SA is abolished.
Results and Discussion
Development of an SA/JA cross-talk mutant screen.
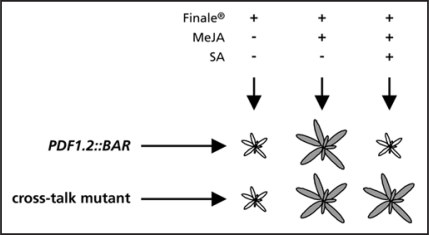
The SA/JA antagonism is manifested when Arabidopsis plants are exposed simultaneously to SA and methyl jasmonate (MeJA). Whereas MeJA treatment alone induces JA-responsive genes, such as PDF1.2, the combined treatment of SA and MeJA results in strong suppression of these genes.15 This well-characterized antagonism was exploited to develop a mutant screen based on selection for herbicide resistance. We devised a construct of the BIALOPHOS RESISTANCE (BAR) gene under transcriptional control of the JA-responsive PDF1.2 promoter. The BAR gene encodes a phosphinothricin acetyltransferase, which detoxifies phosphinothricin (PPT), the active ingredient in the herbicide Finale®. By placing the herbicide resistance gene under the control of the JA-responsive promoter, one should be able to control plant survival by applying Finale®, with or without MeJA. However, inclusion of SA in the chemical treatment will suppress MeJA-induced activation of the PDF1.2 promoter and, thus, these plants will retain their sensitivity to Finale®. Hence, treatment of PDF1.2::BAR plants with a mixture of MeJA and SA, followed by application of Finale® will be lethal as a result of the cross-talk between SA and JA signaling. Ethyl methane sulfonate (EMS) mutagenesis of this transgenic line and screening of the M2 population after MeJA and SA and Finale® treatment should yield cross-talk mutants that are resistant to the herbicide, indicating loss of SA/JA cross-talk (Fig. 1).
Figure 1.
Experimental set-up for an SA/JA cross-talk mutant screen. Transgenic PDF1.2::BAR plants survive Finale® treatment when the BAR resistance gene under control of the PDF1.2 promoter is induced by MeJA, but die after combined treatment with SA, MeJA and Finale® due to SA/JA cross-talk. Conversely, a cross-talk mutant disrupted in the suppression of JA signaling by SA survives the combined treatment of SA, MeJA and Finale®.
SA-mediated suppression of BAR transcription does not lead to loss of herbicide resistance.
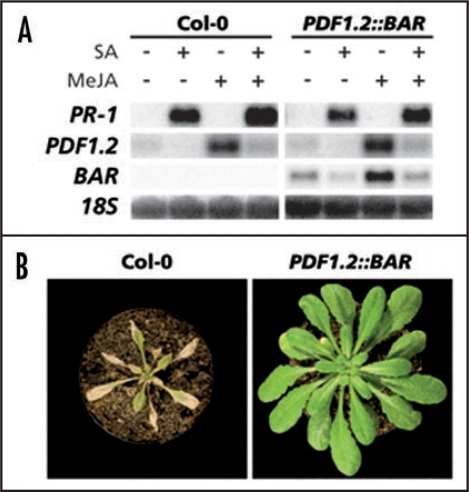
We selected single-insertion PDF1.2::BAR lines and tested them for SA/JA cross-talk by a foliar drench with SA, MeJA or a combination of both chemicals. One line consistently showing SA/JA cross-talk of the PDF1.2::BAR transgene was selected for further study. Figure 2A shows that the SA-responsive PR-1 and JA-responsive PDF1.2 marker genes were induced in this line by SA and MeJA treatment, respectively. The BAR gene was induced by MeJA treatment and suppressed by additional treatment with SA, similar to the endogenous PDF1.2 gene, except for a slightly higher basal expression. We tested whether this basal BAR expression affected herbicide resistance in this line, and found that the PDF1.2::BAR line was already fully resistant to Finale® in the absence of MeJA (Fig. 2B). Application of SA suppressed the basal BAR expression (Fig. 2A). Thus, a combined treatment of SA and Finale® could be expected to be sufficient to kill these transgenic plants in the absence of the PDF1.2-inducer MeJA. This would simplify the mutant selection procedure by eliminating one component necessary to perform the mutant screen.
Figure 2.
SA- and MeJA-responsive gene expression and herbicide tolerance in PDF1.2::BAR. (A) PR-1, PDF1.2 and BAR expression in Col-0 and PDF1.2::BAR plants after foliar drench with 1 mM SA, 0.1 mM MeJA or a combination of both chemicals. Leaf tissue was harvested 24 h after chemical treatment. Equal loading of RNA samples was checked using a probe for 18S rRNA. (B) Plant survival after Finale® treatment of 5-week-old Col-0 and PDF1.2::BAR plants. Photographs were taken 1 week after herbicide treatment.
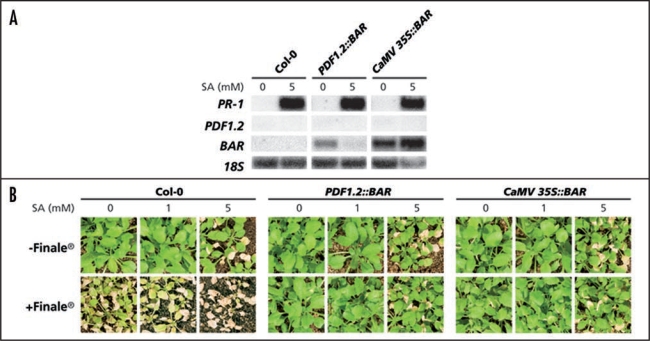
To determine the experimental conditions for a mutant screen, PDF1.2::BAR, Col-0 and CaMV 35S::BAR seedlings were grown on potting soil for 3 weeks before receiving a 10-mL soil drench with 0, 1 or 5 mM SA. One day later, leaf tissue samples were harvested from the 0 and 5 mM SA-treated plants to assess marker gene expression, and the remaining plants were sprayed with Finale®. Plant survival over the course of the next few days was recorded. The gene expression patterns of PR-1, PDF1.2 and BAR in all three genotypes are shown in Figure 3A. The soil drench with SA strongly induced the SA-responsive marker gene PR-1 in all three genotypes. PDF1.2 was not induced by any of the treatments, showing that the constitutive BAR gene expression in the PDF1.2::BAR line was not due to any JA-related stress response. BAR expression in the PDF1.2::BAR line was completely suppressed by the application of SA (Fig. 3A). Thus, the induction conditions were successful in suppressing BAR expression in the PDF1.2::BAR line. However, suppression of PDF1.2::BAR by SA did not decrease the frequency of plant survival. As shown in Figure 3B, most Col-0 control plants had died from the herbicide treatment 5 days after Finale® application, and application of SA had an additive toxic effect. As expected, Finale® had no harmful effect on the constitutive CaMV 35S::BAR line in any of the treatments. Unfortunately, the PDF1.2::BAR line was not sensitive to Finale® either, not even after application of 5 mM SA, which strongly suppressed the expression of PDF1.2::BAR. It seems, therefore, that even very low levels of BAR expression are sufficient to confer herbicide resistance. A similar observation was made in transgenic tobacco lines that expressed the BAR gene under the control of the constitutive CaMV 35S promoter.25 The expression of the BAR gene varied significantly between independent transgenic lines, but lines with even the lowest expression levels were fully resistant to herbicide treatment.25 As the strong reduction in BAR gene expression in response to SA in the PDF1.2::BAR line was not accompanied by a reduction in plant survival, this PDF1.2::BAR line proved unsuitable for the desired SA/JA cross-talk mutant screen.
Figure 3.
Soil-grown PDF1.2::BAR plants survive SA and Finale® treatment. (A) PR-1, PDF1.2 and BAR expression in Col-0, PDF1.2::BAR and CaMV 35S::BAR plants after a soil drench with 0 or 5 mM SA. Leaf tissue was harvested 24 h after chemical treatment. Equal loading of RNA samples was checked using a probe for 18S rRNA. (B) Survival of Col-0, PDF1.2::BAR and CaMV 35S::BAR plants after a soil drench with 0, 1 or 5 mM SA and treatment with Finale® 1 day later. Photographs were taken 5 days after Finale® treatment.
Development of an alternative mutant screening strategy.
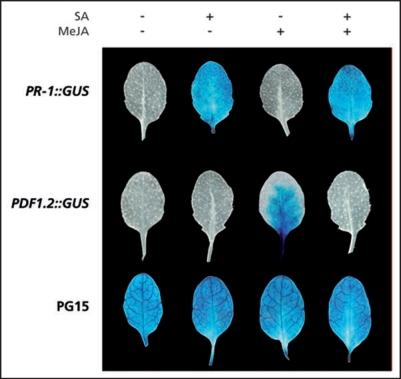
A genetic screen based on a reporter system that allows detection of a quantitative trait might be more successful for the identification of SA/JA cross-talk mutants. The β-glucuronidase (GUS) reporter system allows relatively quick screening of gene expression patterns by histochemical staining of plant tissue. EMS mutagenesis of a PDF1.2::GUS reporter line would be expected to yield putative SA/JA cross-talk mutants, which are characterized by a blue staining, even after combined treatment with SA and MeJA. This screening method is more laborious than the herbicide-based mutant screen, but allows detection of quantitative differences in gene expression. An experimental set-up is shown in Figure 4. Soil drench with a control solution, 1 mM SA, 0.1 mM MeJA or a combination of SA and MeJA was provided to induce SA and JA responses, and trigger cross-talk, respectively. As control lines, the constitutive GUS-expressor PG15 and SA-responsive PR-1::GUS were included. The PR-1::GUS line was induced only in the treatments involving SA, and the MeJA-inducible line PDF1.2::GUS showed clear blue staining upon MeJA treatment, which was completely suppressed upon simultaneous application of SA and MeJA. Hence, application of this approach should lead to a workable screening method for the identification of SA/JA cross-talk mutants.
Figure 4.
GUS activity in PDF1.2::GUS and PR-1::GUS plants. Histochemical staining for GUS expression in 4-week-old PR-1::GUS, PDF1.2::GUS, and the constitutive GUS-expressor PG15 after soil drench with water, 1 mM SA, 0.1 mM MeJA or a combination of both chemicals. Leaf tissue was harvested after 24 h and immersed in GUS staining solution.
Materials and Methods
Cultivation of plants.
Seeds of Arabidopsis thaliana transgenic lines PDF1.2::GUS and PR-1::GUS were kindly provided by Johan Memelink (Leiden University, The Netherlands) and Julia Plotnikova (Massachusetts General Hospital, Boston, USA), respectively. Wild-type accession Col-0, PDF1.2::BAR, CaMV 35S::BAR, PDF1.2::GUS, PR-1::GUS and constitutive β-glucuronidase (GUS)-expressing PG15 plants were grown as described previously.15
Cloning of the BAR construct and plant transformation.
The 1.2-kb fragment of the PDF1.2 promoter was amplified by PCR from genomic DNA of Col-0 plants using the PDF1.2 Fw (5′ GCG AAT TCA TGC ATG CAT CGC CGC ATC G 3′) and PDF1.2 Rv (5′ CGC TCG AGA TGA TTA TTA CTA TTT TGT TTT C 3′) primers, with added EcoRI and XhoI recognition sequences at the 5′ and 3′ ends, respectively. The PDF1.2-promoter fragment was cloned into the XhoI and EcoRI sites of the binary vector pCAMBIA 3200. Using partial digestion with XhoI and full digestion with EcoRI, the pre-existing CaMV 35S-promoter fragment was deleted and replaced by the PDF1.2-promoter fragment. After sequence verification, the binary vector was transformed into Agrobacterium tumefaciens strain AGLO. Col-0 plants were transformed using the floral dip method as described by Clough and Bent,22 and surface-sterilized seeds of transformants were selected on MS medium supplemented with 20 µM MeJA (Serva, Brunschwig Chemie, Amsterdam, The Netherlands) and 20 mg/L DL-phosphinothricin (PPT; Duchefa Biochemie, Haarlem, The Netherlands).
Induction treatments.
Induction treatments for SA/JA cross-talk were performed by foliar drenching with 1 mM SA, 0.1 mM MeJA or a combination of both chemicals, as described previously.15 For herbicide resistance selection conditions, 3-week-old plants received a 10-mL soil drench with 0, 1 or 5 mM SA (Mallinckrodt Baker, Deventer, The Netherlands) and were sprayed the next day with an aqueous solution containing 0.015% (v/v) Silwet L-77 with or without Finale® SL14 (Bayer Cropscience BV, Mijdrecht, The Netherlands) (150 mg/L PPT). For assessing GUS activity, 4-week-old plants received a soil drench with water, 1 mM SA, 0.1 mM MeJA or a combination of both chemicals. The following day, leaves were harvested and assayed for GUS activity by histochemical staining.
Northern blot analysis and GUS assays.
Total RNA was extracted as described previously.23 Northern blots were hybridized with gene-specific probes for PR-1, PDF1.2 and BAR, as described previously.24 The AGI numbers for the genes studied are At2g14610 (PR-1) and At5g44420 (PDF1.2). GUS activity was assessed by transferring a single leaf from individual seedlings to a GUS staining solution as described previously.15
Acknowledgements
We thank Hans van Pelt and Ido Vlaardingerbroek for technical assistance. This research was supported by grants 813.06.002 and 865.04.002 of the Earth and Life Sciences Foundation (ALW), which is subsidized by The Netherlands Organization of Scientific Research (NWO).
Abbreviations
- BAR
bialophos resistance
- CaMV
cauliflower mosaic virus
- EMS
ethyl methane sulfonate
- ET
ethylene
- GRX480
glutaredoxin 480
- GUS
β-glucuronidase
- JA
jasmonic acid
- MeJA
methyl jasmonate
- NPR1
nonexpressor of PR genes 1
- PDF1.2
plant defensin 1.2
- PPT
phosphinothricin
- PR-1
pathogenesis-related 1
- SA
salicylic acid
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/6151
References
- 1.Somerville C, Koornneef M. A fortunate choice: the history of Arabidopsis as a model plant. Nature Rev Genet. 2002;3:883–889. doi: 10.1038/nrg927. [DOI] [PubMed] [Google Scholar]
- 2.Grant MR, Lamb C. Systemic immunity. Curr Opin Plant Biol. 2006;9:414–420. doi: 10.1016/j.pbi.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Howe GA. Jasmonates as signals in the wound response. J Plant Growth Regul. 2004;23:223–237. [Google Scholar]
- 4.Pozo MJ, Van Loon LC, Pieterse CMJ. Jasmonates—signals in plant-microbe interactions. J Plant Growth Regul. 2004;23:211–222. [Google Scholar]
- 5.Van Loon LC, Geraats BPJ, Linthorst HJM. Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 2006;11:184–191. doi: 10.1016/j.tplants.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Von Dahl CC, Baldwin IT. Deciphering the role of ethylene in plant-herbivore interactions. J Plant Growth Regul. 2007;26:201–209. [Google Scholar]
- 7.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 8.Kessler A, Baldwin IT. Plant responses to insect herbivory: The emerging molecular analysis. Annu Rev Plant Biol. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- 9.Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol. 2001;13:63–68. doi: 10.1016/s0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 10.Pieterse CMJ, Dicke M. Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends Plant Sci. 2007;12:564–569. doi: 10.1016/j.tplants.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Beckers GJM, Spoel SH. Fine-tuning plant defence signalling: Salicylate versus jasmonate. Plant Biol. 2006;8:1–10. doi: 10.1055/s-2005-872705. [DOI] [PubMed] [Google Scholar]
- 12.Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol. 2005;43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- 13.Koornneef A, Pieterse CMJ. Cross talk in defense signaling. Plant Physiol. 2008;146:839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spoel SH, Johnson JS, Dong X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA. 2007;104:18842–18847. doi: 10.1073/pnas.0708139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K, Van Loon LC, Dong X, Pieterse CMJ. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell. 2003;15:760–770. doi: 10.1105/tpc.009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50:128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- 19.Dong X. Genetic dissection of systemic acquired resistance. Curr Opin Plant Biol. 2001;4:309–314. doi: 10.1016/s1369-5266(00)00178-3. [DOI] [PubMed] [Google Scholar]
- 20.Pieterse CMJ, Van Wees SCM, Ton J, Van Pelt JA, Van Loon LC. Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biol. 2002;4:535–544. [Google Scholar]
- 21.Glazebrook J. Genes controlling expression of defense responses in Arabidopsis - 2001 status. Curr Opin Plant Biol. 2001;4:301–308. doi: 10.1016/s1369-5266(00)00177-1. [DOI] [PubMed] [Google Scholar]
- 22.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 23.De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC, Dicke M, Pieterse CMJ. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant-Microbe Interact. 2005;18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- 24.Pieterse CMJ, Van Wees SCM, Van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, Van Loon LC. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Block M, Botterman J, Vandewiele M, Dockx J, Thoen C, Gosselè V, Movva NR, Thompson C, Van Montagu M, Leemans J. Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 1987;6:2513–2518. doi: 10.1002/j.1460-2075.1987.tb02537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]