Abstract
The uncontrolled formation of amyloid fibers is the hallmark of more than twenty human diseases. In contrast to disease-associated amyloids, which are the products of protein misfolding, E. coli assembles functional amyloid fibers called curli on its surface using an elegant biogenesis machine. Composed of a major subunit, CsgA, and a minor subunit, CsgB, curli play important roles in host cell adhesion, long-term survival and other bacterial community behaviors. Assembly of curli fibers is a template-directed conversion process where membrane-tethered CsgB initiates CsgA polymerization. The CsgA amyloid core is composed of five imperfect repeating units. In a series of in vivo and in vitro experiments, we determined the sequence and structural determinants that guide the initiation and propagation of CsgA polymers. The CsgA N- and C-terminal repeating units govern its polymerization and responsiveness to CsgB. Specifically, conserved glutamine and asparagine residues present in the CsgA N- and C-terminal repeating units are required for CsgB-mediated nucleation and efficient self-assembly.
Key words: amyloid, nucleation, polymerization, curli, sequence determinants
Bacterial Amyloid Nucleation was Controlled by N- and C-terminal Repeats of CsgA
Amyloid propagation underlies diverse mammalian ailments such as Alzheimer's disease, systemic amyloidosis, bovine spongiform encephalopathy (mad cow disease) and Creutzfeldt-Jacob disease.1 The product of protein misfolding, disease-associated amyloid formation can be erratic and apparently uncontrolled. Certainly, the physiological and physical constraints on disease-associated amyloidogenesis are poorly described and understood. This is due, in part, to the sporadic nature of in vivo amyloid model systems. However, certain organisms have developed the ability to faithfully direct amyloid formation temporally and spatially.2–4 These “functional amyloids” are not toxic to the organism that produces them, but instead fulfill a variety of important physiological roles.2–4 Functional amyloids have been described in bacteria, fungi and mammals.2,3 Because functional amyloid formation can be a tightly controlled process, these biogenesis systems represent a unique platform from which to better understand amyloidogenesis and the cellular toxicity associated with it.
One of the better understood functional amyloids are curli, bacterially produced extracellular fibers required for biofilm and other community behaviors.5 Curli fibers are composed of a major subunit, CsgA, and minor subunit, CsgB. Curli assembly is initiated by the CsgB nucleator protein, which is proposed to provide an amyloid template to CsgA on the cell surface.6 Curli formation in E. coli requires a specific secretion machinery that directs curli subunits to the cell surface and prevents their polymerization inside the cell.5,7,8
At the cell surface, CsgA is secreted as a soluble, unstructured protein that is converted to an amyloid fiber in a CsgB-dependent fashion.9 In vitro, certain CsgB truncation mutants readily assemble into amyloid, which suggested that CsgB might initiate in vivo CsgA amyloid formation in a mechanism akin to seeding.6 Subsequent experiments proved that CsgB-derived amyloids could promote CsgA polymerization in vitro (in a process called heteronucleation).6,10 Preformed CsgA amyloids also promote CsgA polymerization (homonucleation/self-seeding), demonstrating CsgA responds to self-seeding and CsgB nucleation.6,11 To explore the mechanistic details of CsgA polymerization into an amyloid, we extensively analyzed the sequence determinants for curli assembly using a powerful blend of genetic and biochemical approaches.10
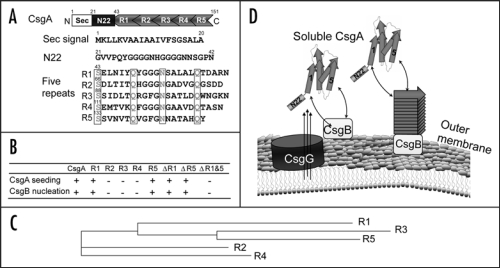
The CsgA amino acid sequence can be divided into three domains, an N-terminal Sec signal sequence (cleaved after secretion through the inner membrane), an N-terminal 22 residues involved in secretion through the outer membrane and five imperfect repeating units (R1, R2, R3, R4 and R5) that comprise the protease-resistant amyloid core (Fig. 1A). We previously showed that peptides representing R1, R3 and R5 are amyloidogenic in vitro.11 Peptides corresponding to R1 and R5 were responsive to CsgA seeding and CsgB nucleation (Fig. 1B).10 Moreover, deletion of R1, R5 or both R1 and R5 resulted in a molecule that no longer polymerized in vivo. A mutant CsgA molecule missing R1 and R5 (ΔR1&R5) was completely incapable of being nucleated by CsgB or being seeded by preformed CsgA fibers (Fig. 1B).10 Interestingly, although the seeding and nucleation competence was determined by R1 and R5, the sequence similarity and identity between of R3 and R5 are higher than those between R1 and R5 (Fig. 1C). It seems that the nucleation/seeding specificity is not simply correlated to sequence similarity or identity. The molecular determinants of this specificity will be an important next step to understand how nature evolved this elegant amyloid propagation system.
Figure 1.
R1 and R5 play important roles in guiding CsgB-responsiveness and CsgA seeding. (A) A schematic of the CsgA primary structure, including a Sec signal sequence (which is cleaved after translocation across the cytoplasmic membrane), N-terminal 22 residues involved in secretion through the outer membrane and five repeating units comprising a protease-resistant amyloid core. The regularly spaced Ser, Gln and Asn residues in each repeating unit are enclosed in boxes. (B) A summary of the interactions between CsgA and CsgB (heteronucleation), or CsgA derivatives (seeding). A “+” means that the interaction reduces the CsgA lag phase. A “−” means that the interaction has little or no affect on the CsgA lag phase. CsgA seeding was measured as previously described11 and CsgB heteronucleation was measured by the overlay assay.10 (C) Phylogram of consensus sequences (SerX5GlnX4AsnX5Gln) of five repeating units. The sequence similarity and identity between consensus sequence of R3 and R5 are higher than those between R1 and R5. (D) The nucleation model of CsgA in vivo polymerization. Curli assembly is governed by surface-localized CsgB (heteronucleation) and by the growing fiber tip (homonucleation). CsgG is the major component of the curli secretion apparatus that directs CsgA and CsgB across the outer membrane. After secretion from the periplasm to extracellular space, R1 and R5 of CsgA can interact with CsgB or fiber tips, initiating curli formation.
From this analysis a simple model of curli assembly can be proposed. After secretion across the outer membrane, the R1 and R5 regions of CsgA would interact with either membrane-associated CsgB, or CsgA fiber tips, which template a conformational change in CsgA so that it now adopts an amyloid form (Fig. 1D).10 The redundancy of seeding/nucleation responsive domains would make curli assembly a highly efficient process.
Bacterial Nucleation Promiscuity Potentially Facilitates Amyloid Propagation
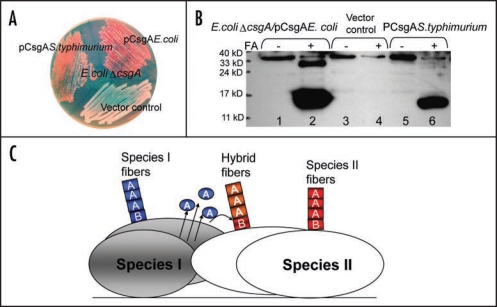
It was reported that seeding effects are strongly influenced by sequence similarity between seeds and soluble amyloid proteins.12–14 For yeast prions, cross-species transmission was determined by amino acid sequence and conformations of prion proteins.15 Prion conversion, including mammal and yeast prions, requires a very high level of identity of the interacting protein sequences.16,17 In contrast to prion proteins, R1 and R5 constitute the critical seeding/nucleation regions of CsgA, although they share only 39% sequence identity between them. In addition, CsgB shares less than 30% sequence identity to CsgA, and it can nucleate CsgA polymerization. The somewhat relaxed requirement of sequence identity for cross-seeding and nucleation in curli biogenesis may have important physiological consequences. It was reported that amyloid-like structures are very abundant in natural biofilms comprising different bacterial species.18 Since the nucleated propagation occurs on the cell surface, bacterial cross-species nucleated amyloidogenesis might happen. Salmonella typhimurium csgA and csgB genes can complement Escherichia coli csgA and csgB mutations in terms of positive Congo red binding, indicating amyloid-like structure formed on the cell surface (Fig. 2A).19 Like E. coli csgA, S. typhimurium CsgA formed SDS-insoluble, fibrous structures in E. coli ΔcsgA cells with the plasmid encoding S. typhimurium CsgA as detected by western analysis and electron microscopy (Fig 2B and data not shown). Presumably, E. coli CsgB converts soluble S. typhimurium CsgA to the amyloid conformation. This cross-species nucleation promiscuity supports the possibility that the CsgA-like molecules of one bacterial species could interact with the CsgB-like molecules of another bacterial species, facilitating the building of complicated matrixes covering bacterial communities (Fig. 2C).
Figure 2.
Salmonella typhimurium CsgA responds to Escherichia coli CsgB in E. coli ΔcsgA cells and assembles into curli. (A) The YESCA plates supplemented with Congo red dye with E. coli ΔcsgA cells transformed with vector control or plasmids encoding E. coli CsgA or S. typhimurium CsgA. Cells were grown for 48 hrs at 26°C. (B) The Western blot of E. coli ΔcsgA cells transformed with vector control (lanes 3 and 4) or plasmids encoding E. coli CsgA (lanes 1 and 2) or S. typhimurium CsgA (lanes 5 and 6). Samples were treated with (+) or without (−) formic acid (FA) to depolymerize the polymers. The blot was probed with anti-CsgA antibody. (C) Schematic shows cross nucleation and polymerization occurring between different bacterial species in the same environment. CsgA molecules secreted from species I (indicated in blue) interact with CsgB of Species II (indicated in red), undergo a conformation change and assemble into hybrid fibers (indicated in orange). Species I and II can assemble their own fibers (indicated in blue and red, respectively).
Remarkably Stringent Requirements of Specific Side-chain Interactions for Curli Assembly
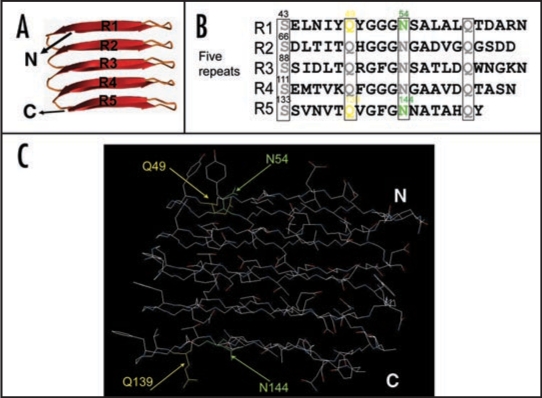
Many proteins, if not all, can assemble into amyloid-like fibers in vitro, which has lead to the suggestion that amyloid formation is an inherent property of polypeptides.1 Clearly, though, the chemical nature of particular side chains in the polypeptide can influence fiber assembly.1 However, the specific role of side-chain interactions during amyloid formation remains poorly described. We addressed this question by dissecting the internally conserved residues in the CsgA repeating units which are predicted to form cross-beta structure (Fig. 3A).20 The consensus sequence (Ser-X5-Gln-X4-Asn-X5-Gln) is found in each CsgA imperfect repeating unit (Fig. 3B). The high degree of the conservation of these polar residues suggests their potential important role in curli assembly. To explore the specific roles of side chains in curli assembly, we analyzed the contribution of conserved polar residues such as Ser, Gln and Asn residues and all the aromatic residues by Ala substitutions (Fig. 3B). We identified four critical residues, Gln and Asn residues at positions 49, 54, 139 and 144 (Fig. 3B and C).21 Ala substitutions of these four residues resulted in a complete loss to CsgB nucleation and an extremely long lag time before fiber formation is initiated, suggesting these four polar side chains contribute to CsgB nucleation response and self-polymerization.21 Surprisingly, Gln residues at position 49 and 139 cannot even be replaced by Asn residues without interfering with curli assembly.21 CsgAQ49N is defective in CsgB nucleation response and self-assembly compared to wild-type CsgA.21 Our findings suggest interaction of CsgA to itself and to CsgB is tightly controlled by very specific side-chain contacts. In addition, we found wild-type CsgA fibers still was able to seed the CsgA mutant with all four critical polar side chains abolished, suggesting CsgA fiber seeding and CsgB nucleation have distinguishable mechanisms.21 It is plausible that fiber mediated seeding is not dependent on polar side chains, whereas the CsgB nucleation requires the participation of polar side chains. All aromatic residues of CsgA do not positively contribute curli assembly.21
Figure 3.
Critical side chains in curli assembly. (A) The predicted structure of CsgA in curli structure. The structure was constructed by Dr. Craig Smith based on previous work from the Kay lab.20 (B and C) The regularly spaced Ser, Gln and Asn residues in repeating units are enclosed in four boxes and their roles in curli assembly were analyzed by Ala substitutions. The critical residues Q49, N54, Q139 and N144 were highlighted in yellow (for Gln residues) and green (for Asn residues) in the primary structure of CsgA (B) and in the predicted 3D structure of five repeating units (C).
Maintenance of native globular structures depends on specific side-chain contacts. Our work demonstrates that the formation of amyloid structure of CsgA also depends on specific side-chain interactions. These side-chain interactions between CsgA-CsgA and CsgA-CsgB allow bacterial amyloid propagation to occur at the correct time and correct location.
Acknowledgements
This work was supported by NIH award AI073847-01.
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/6746
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Hammer ND, Wang X, McGuffie BA, Chapman MR. Amyloids: friend or foe? J Alzheimers Dis. 2008;13:407–419. doi: 10.3233/jad-2008-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid—from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 5.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci USA. 2007;104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem. 2008;283:21530–21539. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Nuallain B, Williams AD, Westermark P, Wetzel R. Seeding specificity in amyloid growth induced by heterologous fibrils. J Biol Chem. 2004;279:17490–17499. doi: 10.1074/jbc.M311300200. [DOI] [PubMed] [Google Scholar]
- 13.Chien P, DePace AH, Collins SR, Weissman JS. Generation of prion transmission barriers by mutational control of amyloid conformations. Nature. 2003;424:948–951. doi: 10.1038/nature01894. [DOI] [PubMed] [Google Scholar]
- 14.Krebs MR, Morozova-Roche LA, Daniel K, Robinson CV, Dobson CM. Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci. 2004;13:1933–1938. doi: 10.1110/ps.04707004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka M, Chien P, Yonekura K, Weissman JS. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B, Newnam GP, Chernoff YO. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc Natl Acad Sci USA. 2007;104:2791–2796. doi: 10.1073/pnas.0611158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 19.Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collinson SK, Parker JM, Hodges RS, Kay WW. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J Mol Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Chapman MR. Sequence Determinants of Bacterial amyloid formation. J Mol Biol. 2008;380:570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]