Abstract
In order to obtain RNA aptamers against bovine prion protein (bPrP), we carried out in vitro selection from RNA pools containing a 55-nucleotide randomized region to target recombinant bPrP. Most of obtained aptamers contained conserved GGA tandem repeats (GGA)4 and aptamer #1 (apt #1) showed a high affinity for both bPrP and its β isoform (bPrP-β). The sequence of apt #1 suggested that it would have a G-quadruplex structure, which was confirmed using CD spectra in titration with KCl. A mutagenic study of this conserved region, and competitive assays, showed that the conserved (GGA)4 sequence is important for specific binding to bPrP and bPrP-β. Following 5′-biotinylation, aptamer #1 specifically detected PrPc in bovine brain homogenate in a Northwestern blotting assay. Protein deletion mutant analysis showed that the bPrP aptamer binds within 25–131 of the bPrP sequence. Interestingly, the minimized aptamer #1 (17 nt) showed greater binding to bPrP and bPrP-β as compared to apt #1. This minimized form of aptamer #1 may therefore be useful in the detection of bPrP, diagnosis of prion disease, enrichment of bPr and ultimately in gaining a better understanding of prion diseases.
Key words: RNA aptamer, prion protein, SELEX, GGA repeat, G-quadruplex
Introduction
The prion protein (PrP) has two alternative forms: a normal cellular protein (PrPC), which is a soluble α-helix rich isoform, and an insoluble β-sheet rich abnormal isoform1 known as the protease-resistant form (PrPSc).2 The detailed mechanism of the structural conversion between the soluble and insoluble forms remains unknown. PrPC is almost ubiquitously expressed and highly conserved in mammals, being anchored on the surface of cells. To gain a better insight into its putative role, much research has gone towards the discovery of molecules that bind prion protein. Of those identified, the most notable are divalent metal ions, several proteins and nucleic acids.3 Knowledge of the function of PrP would contribute towards a better understanding of the processes involved in both the amplification of the infectious agent and the neuronal damage leading to the neuro-degeneration observed in prion diseases. As concerns about the transmission of prion disease in the fields of medicine and food safety increase, there is an increasing demand for an understanding of the processes by which PrPSc can be detected and addressed.4,5
Specific anti-PrP probes, which have high specificity and sensitivity, are required for the diagnosis of prion diseases. Clarification of the binding mechanism between PrP and such probes will assist structural study of PrP. Aptamers are artificial nucleotides derived from systematic evolution of ligands by exponential enrichment (SELEX),6,7 which bind a wide range of targets with a high affinity and specificity, as is seen with antibodies.8,9 Using this method, many aptamers specific to PrP10–20 and PrPSc,12,13 have been isolated.
Recently we have performed an in vitro selection for mouse PrP using an RNA pool with a 30 nt randomized region.16 To prepare a repertoire of anti-PrP RNA aptamers, we carried out SELEX for recombinant bPrP and analyzed for bPrP and bPrP-β. The obtained major RNA aptamers bind to both bPrP and bPrP-β. They possessed tandem GGA repeats (GGA)4 as a consensus sequence, and suggested to form a parallel G-quadruplex structure in the presence of K+ ions. We identified that this core sequence is important for specific binding to bovine PrPs and increased the affinity and selectivity by redesigning this core sequence.
Results
In vitro selection.
To obtain RNA aptamers targeting recombinant bPrP, we carried out SELEX using 97-nt RNA pool that has 55-nt randomized sequences. To increase the selection stringency, we applied the following selection pressures (Table 1): (1) decreased protein concentration and reaction time, and increased washing volumes; (2) increased tRNA concentration as a non-specific competitor; and (3) increased concentration of anti-mPrP RNA aptamer16 as a specific competitor. Furthermore we carried out mutagenic PCR21 to introduce mutations into concentrated sequences (see Materials and Methods). As the enrichment of specific binding RNAs for bPrP was observed in the iterative process (data not shown), G10 RNA pool for bPrP was cloned and sequenced. Interestingly, all RNAs contained tandem GGA repeats, mainly four continuous GGA triplet repeats (GGA)4 (shown underlined in Table 2).
Table 1.
In vitro selection conditions for recombinant bPrP
| Round of selection | RNA (µM) | bPrP (µM) | tRNA (µM) | Anti-PrP RNA aptamer16 (µM) | Time (min) |
| 1 | 10 | 2 | 0 | 0 | 60 |
| 2 | 6 | 0.4 | 10 | 0 | 30 |
| 3 | 4 | 0.2 | 40 | 8 | 20 |
| 4 | 3 | 0.1 | 100 | 16 | 15 |
| 5 | 2 | 0.3 | 100 | 32 | 10 |
| 6 | 2 | 0.05 | 100 | 48 | 10 |
| 7 | 2 | 0.05 | *6 | 0 | 10 |
| 8** | 2 | 0.05 | 100 | 64 | 10 |
| 9** | 2 | 0.05 | 100 | 64 | 10 |
| 10 | 6 | 0.05 | 100 | 96 | 10 |
U13 was used instead of tRNA.
The mutagenic PCR was introduced.
Table 2.
Sequences of randomized regions and majorities of isolated RNA aptamers against recombinant bPrP
| Name of clone | Number of clones | Sequence of randomized region |
| #1 | 14/30 | 5′-CAAUCCAUUCAUCUCUCGAAUGAGGAAGUAGCCCAAGAGGAGGAGGAGGAUGAGC-3′ |
| #6 | 11/30 | 5′-ACCUUCUGUUCAUAUCGUGACCAACCCAAUAGAUUGUGAUAAAGGAGGAGGAGGA-3′ |
| #20 | 3/30 | 5′-UUGCCAAUAGCCCUACGGAGGAGGAGGCUGGACUAUAGACAGUUUACUUAAUAAA-3′ |
| others | 5′-CGCUCUGCCUGCGACCCAUCGACGUGAGUUUGCGAAAAAAGGAGGAGGAGGA-3′ | |
| 5′-UAGCCCCCCUAGGAGCCGCUUGUGAUAAGAGUUCAAAUUGGAGGAGGUGGAUUGA-3′ |
The conserved GGA triplet repeats (GGA)4 are underlined involving its one point mutations.
Binding affinities of apt #1 against bPrP or bPrP-β.
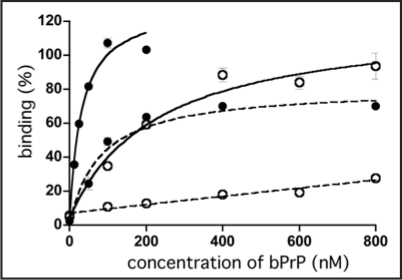
The representative RNA aptamers designated apt #1 and apt #6 were analyzed. To determine their respective Kd values, a filter binding assay was performed using 10 nM RNA and various concentrations of bPrP or bPrP-β in selection buffer [20 mM Tris-HCl (pH 7.5), 100 mM NaCl]. Apt #1 showed a higher affinity (Fig. 1, Kd = 82 ± 21 nM for bPrP) than that of apt #6 (data not shown, Kd = 166 ± 55 nM for bPrP). Because of the structural difference between bPrP and bPrP-β, binding affinity of both RNA aptamers for bPrP-β are over 10-fold lower than those for bPrP. As it demonstrated a higher affinity than apt #6, we focused on apt #1 in subsequent analyses.
Figure 1.
Binding of apt #1 to bPrP and bPrP-β. Binding curves of apt #1 to bPrP and bPrP-β are shown by closed circles and open circles, respectively. Solid and dashed lines represent different buffer conditions: 10 mM K+ and 100 mM Na+, respectively. The binding data are analyzed by GraphPad PRISM (see Materials and Methods).
The (GGA)4 sequence suggested that it forms a G-quartet structure. The formation of a G-quartet structure is stabilized by monovalent cations such as K+ and Na+ ions.25,26 In general, DNA/RNA G-quadruplexes bind K+ over Na+ ions.25,26 As is clearly shown in Figure 1, binding affinities for both proteins bPrP and bPrP-β in 10 mM K+ ion (Kd = 31 nM and 220 nM, respectively; solid lines) were increased in comparison with 100 mM Na+ ion (dashed lines). We also tested apt #1 binding without salt (Kd = 50 nM for bPrP and ≥1000 nM for bPrP-β; data not shown). The presence of potassium ions conferred a greater affinity of apt #1 to bPrPs, which was particularly profound for bPrP-β. We therefore prepared a buffer solution containing K+ ions to be used in subsequent analyses on apt #1 (20 mM Tris-HCl (pH 7.5), 10 mM KCl).
Detection of bPrPC in bovine brain homogenate using apt #1.
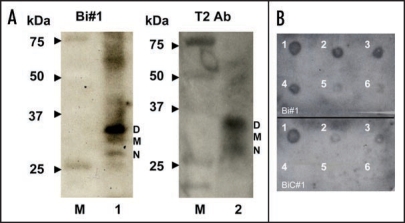
To test the ability of apt #1 to detect bPrPC, we performed Northwestern blotting using a 5′-biotinylated apt #1 (Bi#1) with streptavidin-alkaline phosphatase conjugates (SA-AP) as a secondary probe (left in Fig. 2A). Bi#1 detected the three types of bPrPC (non-, mono- and di-glycosylated forms of PrPC; lane 1) in bovine brain homogenate as well as antibody T2 (lane 2). The epitope of antibody T2 is located between amino acids 147 and 152.23 This result indicates that apt #1 is applicable as a detection tool for bPrP and may perform as well as antibodies.
Figure 2.
Detection of bPrPC in bovine brain homogenate and dot-blotting assay using a biotinylated apt #1 (Bi#1). (A) Bovine brain homogenate was analyzed using SDS-PAGE and subsequently with Northwestern blotting. bPrPC (lane 1 and 2) was detected using Bi#1 (left) and T2 antibody (right). Lane M represents protein molecular weight markers. N, M and D indicate non-, mono- and di-glycosilated PrP isoforms, respectively. (B) Dot-blotting assay of recombinant bPrP using Bi#1 or BiC#1. Number 1 represents cross-linked 5′-biotinylated RNA (Bi#1, BiC#1). Numbers 2–6 represent decreasing concentration of mounted bPrP (ng) as follows: 2 = 480, 3 = 240, 4 = 120, 5 = 60, 6 = 30.
The detection limit of bPrP with Bi#1 was approximately 30 ng (spot 6 in the upper of Fig. 2B) using dot blot analysis in the presence of poly (U) as a non-competitive binder. A negative control BiC#1 that has the complementary sequence of apt #1, showed a detection limit 240 ng (spot 3 in the lower of Fig. 2B), indicating specific binding of apt #1 against bPrPC.
Conserved region (GGA)4 suggests a parallel G-quadruplex.
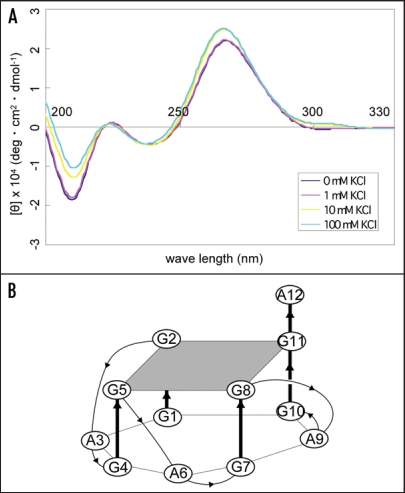
To examine the structure of apt #1, we measured CD spectra of the aptamer in the different concentrations of K+ ion. It is known that a parallel G-quadruplex gives a positive CD peak at 260 nm, while an antiparallel G-quadruplex gives a positive CD peak at 295 nm either with or without a positive CD peak at 260 nm in the presence of monovalent cations.27,28 A peak was observed at 260 nm of CD and the peak intensity increased as the concentration of K+ ions increased, with maximum peak intensity at 10 mM KCl (Fig. 3A). This result suggests that apt #1 forms a parallel G-quadruplex in the presence of K+ ion, with saturation occurring at 10 mM KCl. We also confirmed no increase of peak intensity at 260 nm in the presence of K+ ion on the apt #1 mutant with substitutions of G tracts in the conserved (GGA)4 region (data not shown).
Figure 3.
CD spectra of apt #1 and r(GGA)4-15 in the presence of KCl. CD spectra were measured at 20°C in titration with KCl (0, 1, 10, 100 mM). (A) CD spectra of 10 µM apt #1. (B) The structure of d(GGA)4.24
Matsugami et al. reported that d(GGA)4 folds into an intramolecular G-quadruplex composed of a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad with parallel orientation under physiological K+ conditions24 (Fig. 3B). The CD spectra of apt #1 (Fig. 3A) showed a similar pattern with that of d(GGA)4. Although direct evidence is lacking, we believe that apt #1 also forms the similar type of the parallel G-quadruplex structure. Previous reports have described G-quadruplex forming RNA aptamers for PrP,10,11 but sequences are different from described here.
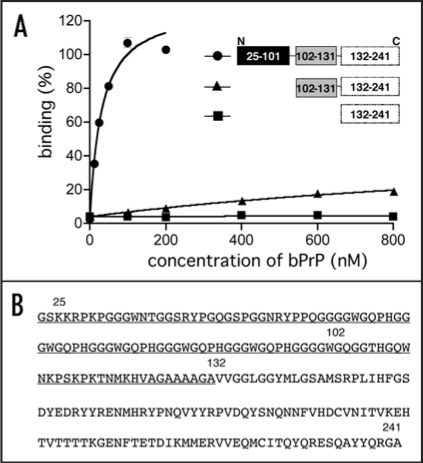
Aptamer binding sites locate amino acids 25–131.
To investigate the apt #1 binding region of bPrP, we performed binding analysis with deleted proteins: bPrP (102–241) and bPrP (132–241) (Fig. 4A). Apt #1 bound strongly to full length bPrP (Kd = 31 nM), while Kd value of the deletion variant bPrP (102–241) (≥1000 nM; indicated with triangles) increased more than 30-fold compared with bPrP. Apt #1 could not bind to variant bPrP (132–241). Consequently, the binding region for apt #1 is located within amino acid residues 25–131, underlined in Figure 4B. Deletion of N-terminal 25–101 residues produced a strong effect for the aptamer binding. This suggests that this region is the main binding site as well as an anti-mPrP aptamer.16 Mercey et al. reported that the two lysine clusters (amino acids 25–32 and 102–110 in ovine PrP) are primary nucleic acids binding sites, the former region showing stronger binding than the latter.15 Considering these results, it seems that apt #1 is also likely to bind to the two lysine clusters, although there is no homology between apt #1 and ovine-PrP aptamers.
Figure 4.
Binding affinity of anti-bPrP aptamer apt #1 for deletion variants of bPrP. (A) Circles, triangles and squares indicate full length bPrP (25–241), bPrP (102–241) and bPrP (132–241), respectively. (B) The amino-acid sequence alignment of bPrP. The apt #1 binding region is underlined.
The conserved region (GGA)4 is important for binding to bPrP or bPrP-β.
To investigate the correlation between binding affinity and G-quadruplex structure of the conserved (GGA)4, we prepared several mutants of the (GGA)4 region (m1–m5) and analyzed their affinities compared to bPrP and bPrP-β. The Kd values for different mutants are shown in Table 3. All #1 mutants showed decrease of binding ability to bPrP-β. The conserved (GGA)4 is therefore important for binding to bPrP-β. In the case of interactions with bPrP, all mutants except m4 showed moderate decrease in binding affinity (1.8–2.5-fold). This is due to the ability of PrP to bind to nucleic acids.29,30
Table 3.
Sequences and binding affinities of different mutants
| aptamers | sequence | Kd (nM) | |
| bPrP | bPrP-β | ||
| apt #1 | 5′-GGAGGAGGAGGA-3′ | 31 ± 6 | 220 ± 67 |
| m1 | 5′-GGAGGAGGA-3′ | 79 ± 24 | ≥1000 |
| m2 | 5′-GGAGGA-3′ | 75 ± 28 | ≥1000 |
| m3 | 5′-(no GGA repeat)-3′ | 55 ± 24 | ≥1000 |
| m4 | 5′-UUUUUUUUUUUU-3′ | 134 ± 42 | ≥1000 |
| m5 | 5′-UUAUUAUUAUUA-3′ | 56 ± 16 | ≥1000 |
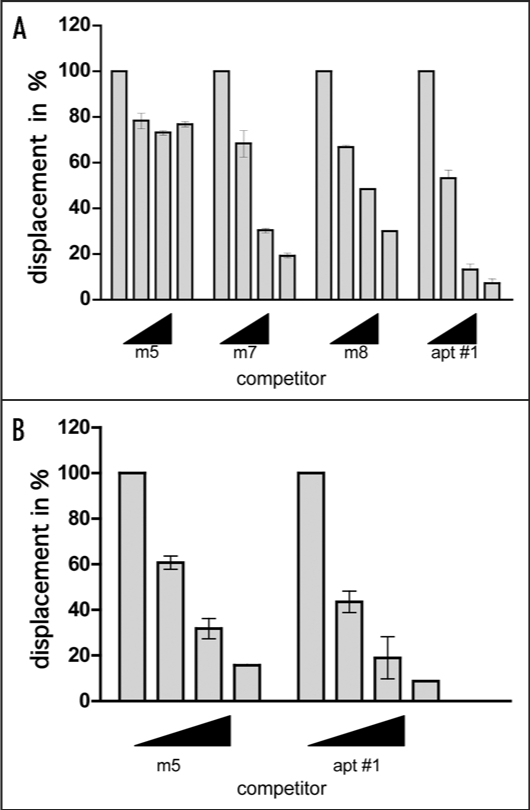
Since we could not verify the specific binding of mutants to bPrP from the determined Kd values, we carried out competitive binding analysis using the labeled apt #1 in the presence of elevated amounts of unlabeled competitors (Fig. 5). We used non G-quadruplex forming competitor (m5) and probable G-quadruplex forming competitors (m7, m8, apt #1). It was demonstrated that apt #1 was the best competitor, and all probable G-quadruplex forming competitors competed with apt #1 for binding to bPrP in dose-dependent manner. There are small differences of displacement efficiency among apt #1, m7 and m8. By comparing the competing ability of G-quadruplex forming RNAs m7, m8 and apt #1, the adenine residues in (GGA)4 are also important for binding to bPrP. Non G-quadruplex forming competitor (m5) showed weak competition in the presence of K+ ion. Under the no salt condition m5 competed with apt #1 in the similar level of apt #1 (Fig. 5B). This means that binding ability of apt #1 to bPrP is specific binding in the presence of K+ ion.
Figure 5.
Comparison of binding of apt #1 and bPrP using competitive binding assay. The #1 mutants (m7, m8) contained substitutions (shown underlined) in the conserved region (GGA)4 as follows: m7: 5′-GGUGGAGGAGGA-3′; m8: 5′-GGUGGUGGUGGU-3′. The competitor RNAs were added in various concentrations with 0, 1, 5 and 10-fold greater molar concentrations of labeled apt #1. Binding assay was performed in the presence of 10 mM KCl (A) and in the absence of KCl (B).
Minimization of apt #1 retained the binding ability to bPrP and bPrP-β.
Since the conserved region (GGA)4 of apt #1 plays important role for specific binding to bPrP and bPrP-β, we minimized the aptamer to a 12-nt RNA r(GGA)4 and measured Kd values (Table 4). Compared with apt #1, r(GGA)4 showed better affinity for bPrP (Kd = 8.5 nM) and similar Kd value for bPrP-β (Kd = 280 nM). We then examined d(GGA)4, which forms the unique G-quadruplex structure shown in Figure 3B.24 This revealed that d(GGA)4 showed a strikingly lower affinity for both bPrP and bPrP-β, compared to r(GGA)4. This result suggests that r(GGA)4 has a different G-quadruplex structure with that of d(GGA)4.
Table 4.
Comparison of binding affinities for bPrP and bPrP-β between GGA repeat containing RNAs and DNA
| RNA/DNA | sequence | Kd (nM) | |
| bPrP | bPrP-β | ||
| apt #1 | - | 31 ± 6 | 220 ± 70 |
| r(GGA)4 | r(GGAGGAGGAGGA) | 8.5 ± 3.4 | 280 ± 80 |
| d(GGA)4 | d(GGAGGAGGAGGA) | 64 ± 31 | ≥1000 |
| R14 | r(GGAGGUUUUGGAGG) | 32 ± 18 | 610 ± 240 |
To test the RNA sequence specificity of r(GGA)4, we analyzed the binding of R14, which contains GGA residues and forms another type of intramolecular G-quadruplex consisting of tetrad and hexad in the presence of K+.31 As shown in Table 4, R14 showed greater affinity for both proteins compared to d(GGA)4, but lower affinity than r(GGA)4. Hence the conserved tandem GGA repeat (GGA)4 is the important region for binding to bPrP and bPrP-β, and the minimized 12-nt RNA retained this binding ability.
Adenine stretch attached at 5′-site of (GGA)4 enhanced the binding affinity for bPrP-β.
Besides of conserved region (GGA)4, adenine tracts (3–6 residues) were frequently observed at the 5′-site of (GGA)4 in obtained RNA aptamers (Table 2). The capacity of these adenine sequences to bind to bPrPs was investigated. The sequence of constructs and their Kd values are shown in Table 5. Binding affinities were almost the same (Kd = 9–17 nM) for full-length bPrP. For truncated variant bPrP (102–241), they also showed the same level of binding affinities (Kd = 160–270 nM) except r(GGA)4-19 attached 7 adenines (Kd = 540 nM). However with regard to bPrP-β, the five adenine stretch r(GGA)4-17 showed the lowest Kd value (Kd = 78 nM), followed by the four adenines stretch r(GGA)4-16 (Kd = 92 nM). The optimal numbers of adenine correspond with the trend observed in the RNA aptamers.
Table 5.
Comparison of adenine stretch at 5′-site of (GGA)4 with binding affinities of bPrP, bPrP(102–241) and bPrP-β
| aptamers | sequence | Kd (nM) | ||
| bPrP | bPrP (102–241) | bPrP-β | ||
| r(GGA)4-19 | AAAAAAAGGAGGAGGAGGA | 17 ± 5 | 540 ± 310 | 270 ± 90 |
| r(GGA)4-18 | AAAAAAGGAGGAGGAGGA | 10 ± 2 | 160 ± 50 | 150 ± 40 |
| r(GGA)4-17 | AAAAAGGAGGAGGAGGA | 9.4 ± 4.0 | 270 ± 120 | 78 ± 30 |
| r(GGA)4-16 | AAAAGGAGGAGGAGGA | 16 ± 3 | 190 ± 50 | 92 ± 26 |
| r(GGA)4-15 | AAA GGAGGAGGAGGA | 16 ± 4 | 220 ± 90 | 200 ± 90 |
| r(GGA)4 | GGAGGAGGAGGA | 8.5 ± 3.4 | 240 ± 90 | 280 ± 80 |
As reported previously, amino acids 23–90 (human numbering) of PrP interact with nucleic acids in a specific or non-specific manner.32 Short RNAs including r(GGA)4-17 bound to bPrP (102–241) (probably amino acids residues 102–131 from the binding result of #1 in Fig. 4) in a specific manner. Therefore, like an antibody, r(GGA)4-17 shown in Table 5 may recognize PrP27–30, which is a proteinase K (PK) resistant fragment following removal of the N-terminal region (approximately up to amino acid position 90 in mPrP).33–36
Discussion
In this study, we described an in vitro selection of RNA aptamers against recombinant bPrP. All selected RNA aptamers contained the four repeats of a two guanine containing sequence and most of RNAs contained the conserved sequence (GGA)4 (Table 2). Our findings suggest that apt #1 can be used to detect PrPC in bovine brain homogenate, a process which more conventionally employs an antibody with an immuno-blotting assay (Fig. 2). By collecting CD spectra in increasing concentrations of KCl, apt #1 probably forms a parallel G-quadruplex derived from the (GGA)4 sequence.
It has been reported that the proposed G-quadruplex structure plays a critical role in the binding to PrP.10,11 Among the reported anti-PrP RNA aptamers containing GGA repeats, anti-syrian golden hamster PrP aptamers contain GGGA repeat interacting with amino acids 23–52,10 and aptamer DP7 targeted to amino acids 90–129 of human PrP involves GGA repeats.11 Anti-mPrP aptamer 60-3 used as a competitor in this study also contains GGAGG repeat interacting with amino acids 23–119.16 The aptamer 60-3 binds to bPrP in the presence of Na+,16 however it did not show enhanced bindings to both bPrP and bPrP-β in the presence of K+ (data not shown). The conserved sequence of tandem GGA repeat (GGA)4 is isolated only in this study. Considering the high frequent appearance of GGA observed in anti-PrP RNA aptamers, GGA repeat must be meaningful for specific binding to PrPs.
We confirmed that the conserved sequence (GGA)4 is important for specific binding to both bPrP and bPrP-β by investigating the effect of substitutions in this region (Table 3) and competitive binding analysis (Fig. 5). Amongst G-quartets which form small DNA and RNAs involving a GGA repeat, r(GGA)4 showed the best affinity to both bPrP and bPrP-β indicating the minimal sequence required for the sequence specificity for preferential binding (Table 4). d(GGA)4 and R14,31 form similar intramolecular parallel G-quadruplexes consisting of tetrad-heptad and tetrad-hexad, respectively.24 R14 showed better binding to bPrP and bPrP-β compared to d(GGA)4. This preferential binding may be derived either from the RNA molecule or differences in the tertiary G-quadruplex structures. To clarify the relationships between binding and structure, we are in the process of investigating these structures in more detail.
The binding affinity of bPrP-β with the G0 RNA pool was notably low (data not shown). The recombinant amyloidogenic prion protein, bPrP-β, is generated through chemical treatment of recombinant bPrP which enhances the formation of the β-sheet secondary structure in the C-terminal region.37 The N-terminus of PrP (amino acids 23–120 in mPrP) is a flexible non-structural region and the C-terminus of PrP is preserved in the globular three-dimensional structure.38 It is known that the N-terminal region of PrP interacts with nucleic acids in non-specific and/or specific manner.29,30,32 In an NMR study of the interaction between murine recombinant PrP and RNA aptamer SAF93,12,43–59 it was observed that part of the N-terminal domain is modified after interaction with RNA.39 From these experimental results, it can be deduced that the environment of N-terminal regions also differ between bPrP and bPrP-β. One possibility is that the packed structure of the C-terminal region, which contains higher numbers of β-sheets in bPrP-β, causes a narrowing of free space and a decrease in structural flexibility in the N-terminal region, resulting in poor acceptability of nucleic acids and induced fitting. Finally, we showed that the binding ability for bPrP-β could be improved by minimization of apt #1 to a 17 nt RNA containing an adenosine sequence at the 5′-site of (GGA)4.
By investigating the binding properties of deletion variants of bPrP, we identified the N-terminal region of bPrP, amino acids 25–131, as being a key region for apt #1 binding. Apt #1 could bind to amino acids 102–131 of bPrP. It is known that amino acids 25–101 (23–90 in hPrP) contain the nucleic acid binding site.32 Notably, the additional N-terminal binding region 102–131 provides apt #1 with specific binding activity. Furthermore, it conferred the ability of the minimized aptamer to bind with both bPrP and bPrP-β with high affinity.
Aptamers, with their high ligand specificity, can enrich a target molecule from biological samples. For example, RNA ligands have been successfully used for the concentration of PrPC and PK-resistant PrP taken from serum, urine32 and brain homogenate.40 Previously, we also used anti-mPrP aptamer for bead-based purification and concentration of PrPC from mouse brain homogenate.16 Takemura et al. proposed that PrPSc-enrichment with pre-treatment using PrPC-specific aptamers, to remove normal prions from a sample, could be applied as diagnostic tools in double ligand assay systems.17 Such an enrichment method of PrPSc might be required for early stage detection of the β form PrP because it exists in extremely low proportions compared to the α form. After treatment with PrPC-specific ligands, apt #1 may be useful for PrPSc enrichment, rather than employing an antibody without PK treatment.
In a recent publication investigating the interaction between RNAs and recombinant PrP, it was shown that PrP aggregates could be induced by the presence of small synthetic oligonucleotides and aggregates with total RNA extract from cultured mouse neuroblastoma cells (N2a) were cytotoxic.39 A sensitive method of pathological prion protein detection using cyclic amplification of protein misfolding (PMCA) has been developed.41 It has been reported that successful PMCA propagation of PrPSc molecules in a purified system requires accessory poly (A) RNA molecules.42 Association with PrP and some specific RNAs such as RNA aptamers might help in this field.
In conclusion, we succeeded in harvesting an RNA aptamer against bPrP and bPrP-β, and in producing a high affinity, and a 17 mer minimized form containing 5 adenines at the 5′-site of (GGA)4. Its ability to bind to bPrP (102–241), which corresponds to PrP 27–30, indicates that it may detect PK-digested PrP as well as antibodies. These aptamers will, we believe, be valuable in the study of conversions, diagnosis and therapy of prion disease.
Materials and Methods
Proteins.
Recombinant bPrP (amino acids 25–241), amyloidogenic PrP prepared from recombinant bPrP (bPrP-β) and truncated bPrPs [bPrP (102–241): amino acids 102–241; bPrP (132–241): amino acids 132–241] were purchased from Alicon (Switzerland).
In vitro selection of RNA aptamers against bPrP.
An N55S RNA pool containing a region of 55 randomized nucleotide [5′-GGGAGGUGGAACUGAAGGAGA-(N55)-ACUUCGCAAUCGCUCUACGCA-3′] was used to perform in vitro selection on a 0.45 µm HAWP nitrocellulose filter (Millipore). RNAs were heat denatured at 90°C for 2 min, 72°C for 5 min, 55°C for 5 min, 37°C for 2 min and cooled to room temperature in 50 µl of binding buffer [20 mM Tris-HCl (pH 7.5) and 100 mM NaCl] prior to use. The RNA pool was incubated for 10 to 30 min with bPrP in the presence of competitor RNAs (tRNA and/or antimPrP aptamer 60-3,16 at room temperature. This mixed solution was passed through the nitrocellulose filter and washed with the binding buffer. RNA bound to bPrP on the filter was recovered with 400 µl of 7 M urea at 90°C for 5 min. The eluted RNA was ethanol precipitated and reverse transcribed using AMV reverse transcriptase (Wako) at 42°C for 1 h. The product was PCR amplified (94°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec) using Gene Taq (Nippon Gene) with forward primer [5′-TGTAATACGACTCACTATAGGGAGGTGGAACTGAAGGAGA-3′] and reverse primer [5′-TGCGTAGAGCGATTGCGAAGT-3′] (Fasmac), and transcribed using the T7 Ampliscribe Kit (Epicentre Technologies). The RNA product was treated with DNase I and purified by Micro Bio-Spin Columns P-30 (Bio-Rad) or 8% PAGE containing 7 M urea and subjected to the next round of selection. From the eighth round of selection mutagenic PCR21 was introduced. After the tenth generation cDNA pool was inserted into the pGEM-T Easy vector (Promega), cloned in Escherichia coli JM109 strain, and sequenced (ABI 3100; Applied Biosystems).
Preparation of RNA aptamer.
To prepare RNA aptamer, the double-stranded DNA generated by PCR was used as a template for in vitro transcription by T7 RNA polymerase, as described above.
Mutant RNAs of apt #1 were prepared from mutagenic PCR using the following DNA templates (Fasmac) and the selection primers: for m1 [5′-TGC GTA GAG CGA TTG CGA AGT TGC TCA UCC UCC UCC TCT TGG GCT ACT TCC TCA TTC GAG AGA TGA ATG GAT TGT CTC CTT CAG TTC CAC CT CCC-3′]; for m2 [5′-TGC GTA GAG CGA TTG CGA AGT TGC TCA UCC UCC TCT TGG GCT ACT TCC TCA TTC GAG AGA TGA ATG GAT TGT CTC CTT CAG TTC CAC CTC CC-3′]; for m3 [5′-TGC GTA GAG CGA TTG CGA AGT TGC TCA TCT TGG GCT ACT TCC TCA TTC GAG AGA TGA ATG GAT TGT CTC CTT CAG TTC CAC CTC CC-3′]; for m4 [5′-TGC GTA GAG CGA TTG CGA AGT TGC TCA AAA AAA AAA AAA TCT TGG GCT ACT TCC TCA TTC GAG AGA TGA ATG GAT TGT CTC CTT CAG TTC CAC CTC CC-3′]; for m5 [5′-TGC GTA GAG CGA TTG CGA AGT TGC TCA TUU TUU TUU TUU TCT TGG GCT ACT TCC TCA TTC GAG AGA TGA ATG GAT TGT CTC CTT CAG TTC CAC CTC CC-3′]; for m7 [5′-TGC GTA GAG CGA TTG CGA AGT TGC TCA TCC TCC TCC ACC TCT TGG GCT ACT TCC TCA TTC GAG AGA TGA ATG GAT TGT CTC CTT CAG TTC CAC CTC CC-3′]; for m8 [5′-TGC GTA GAG CGA TTG CGA AGT TGC TCA ACC ACC ACC ACC TCT TGG GCT ACT TCC TCA TTC GAG AGA TGA ATG GAT TGT CTC CTT CAG TTC CAC CTC CC-3′]. To prepare C#1, the generated PCR fragment using plasmid #1 as template and the proper primers (+) c#1 and (−) c#1, were used for in vitro transcription by T7 RNA polymerase as described above. (+) c#1: [5′-TGT AAT ACG ACT CAC TAT AGG CGT AGA GCG ATT GCG AAG-3′]; and (−) c#1: [5′-GGG AGG TGG AAC TGA AGG AGA-3′].
The chemically synthesized RNAs or DNAs: r(GGA)4-15, r(GGA)4, d(GGA)4, R14, r(GGA)4-16, r(GGA)4-17, r(GGA)4-18 and r(GGA)4-19 were purchased from Fasmac (Japan). 5′-biotinylated RNAs (Bi#1, BiC#1) were prepared with a 5′ end tag nucleic acid labeling system (Vector Laboratories).
Binding assay of anti-bPrP aptamer.
Radioisotope labeling of RNA by in vitro transcription was carried out using α-32P-ATP, as previously described.16 Refolded 32P labeled aptamer (10 nM) was mixed with varying concentrations of bPrP, or its derivatives, to a total volume of 25 µl in reaction buffer [20 mM Tris-HCl (pH 7.5), 100 mM NaCl or 10 mM KCl]. After 20 min incubation, the mixture was passed through a nitrocellulose filter and washed with 500 µl of the reaction buffer. The amount of bound RNA was measured with BAS 2500 (Fuji Film), and binding activities were calculated as the percentage of input RNA retained on the filter in the protein-RNA complex. We determined the equilibrium dissociation constant (Kd) using GraphPad PRISM using non-linear regression curve fitting, and a one site binding hyperbola equation (RNA binding (%) = Bmax × [PrP]/(Kd + [PrP]), where Bmax is the maximum bound at saturating PrP concentrations).
Northwestern assay with the aptamer.
The Northwestern assay was performed using conventional methods.22 A sample of 10 µl 10% bovine brain homogenate in suspension buffer [0.1% Nonidet P-40, 0.1% deoxycholate, 20 mM Tris-HCl (pH 7.5) and 100 mM NaCl] was separated by 10% SDS-PAGE and transferred to a 0.22 µm nitrocellulose membrane (Bio-Rad). A sample of 2 ml 1% BSA in binding buffer [20 mM Tris-HCl (pH 7.5) and 10 mM KCl] was used for blocking for 30 min. Proceeding this were 30 min incubation, with 200 nM of 5′-biotin labeled apt #1 (Bi#1) in binding buffer, followed by a 30 min incubation with streptavidin-alkaline phosphatase conjugate (SA-AP; 2 ng/µl; Roche Applied Science) in binding buffer and a 1 min incubation with CDP-Star (20 mg/ml, Roche Applied Science). Detection was carried out using ECL Mini-Camera (Amersham Biosciences) and Polaroid film. For the standard immuno-blotting assay of bPrPC, a T2 antibody was used.23 All procedures were performed at room temperature.
Dot-blotting assay.
5′ biotinylated RNA, Bi#1 or BiC#1, was spotted on nitrocellulose membrane and UV cross-linked for 5 min. Different amounts of bPrP (480, 240, 120, 60 and 30 ng) were spotted and air-dried for 10 min. The membrane was treated by blocking with 1 % BSA in reaction buffer [20 mM Tris-HCl (pH 7.5), 10 mM KCl] for 30 min, incubating with 200 nM of Bi#1 or BiC#1 in the presence of poly U (0.5 µg/µl, Amersham Biosciences) for 20 min, and then incubating with SA-AP (2 ng/µl) in reaction buffer for 30 min. Following a 1 min incubation with CDP-Star (20 mg/ml) in 0.1 M Tris-HCl (pH 9.5), detection was carried out with ECL Mini-Camera and Polaroid film. All procedures were performed at room temperature.
Circular dichroism spectroscopy.
CD spectra were recorded with a JASCO J-720 Spectropolarimeter (JASCO) as previously reported.24 A cell of 1 mm light path length and 300 µl volume was used to analyze in titration with KCl (0, 1, 10, 100 mM). The spectra were scanned four times from 200 nm to 330 nm. The CD intensities were expressed in [θ] per residue.
Competitive binding assay against bPrP.
To characterize the specific binding of apt #1, binding of apt #1 to bPrP with increasing concentrations of competitor RNAs or DNAs were measured using a filter-binding assay. The labeled apt #1 (25 nM) was incubated with bPrP (50 nM) in the presence of a competitor (0, 1, 5 and 10-fold molar ratios) in binding buffer [20 mM Tris-HCl (pH 7.5), 10 mM KCl] for 20 min. The mixture was passed through a nitrocellulose filter, washed with 500 µl binding buffer, and the amount of binding of labeled apt #1 was determined as described above. 100% maximal binding is defined as the percentage of apt #1 bound to bPrP in the absence of any competitor.
Acknowledgements
The authors thank Drs. Katahira and Matsugami of Yokohama City University for measurement of CD spectra and helpful discussions. This work was supported by the fund from National Institute of Advanced Industrial Science and Technology (to S.N.) and by Grant-in-Aid from the BSE Control Project of the Ministry of Agriculture, Forestry and Fisheries of Japan (to T.Y.).
Abbreviations
- bPrP
bovine prion protein
- bPrP-β
β isoform of bovine prion protein
- bPrP (102–241)
amino acids 102–241 of bovine prion protein
- bPrP (132–241)
amino acids 132–241 of bovine prion protein
- mPrP
mouse prion protein
- PrPC
cellular isoform of prion protein
- PrPSc
scrapie isoform of prion protein
- SELEX
systematic evolution of ligands by exponential enrichment
- CD
circular dichroism
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/7024
References
- 1.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marc D, Mercey R, Lantier F. Scavenger, transducer, RNA chaperone? What ligands of the prion protein teach us about its function. Cell Mol Life Sci. 2007;64:815–829. doi: 10.1007/s00018-007-6370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wadsworth JD, Collinge J. Update on human prion disease. Biochim Biophys Acta. 2007;1772:598–609. doi: 10.1016/j.bbadis.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Hu W, Kieseier B, Frohman E, Eagar TN, Rosenberg RN, Hartung HP, Stuve O. Prion proteins: physiological functions and role in neurological disorders. J Neurol Sci. 2008;264:1–8. doi: 10.1016/j.jns.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 6.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 7.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, Yang D, Schluesener HJ, Zhang Z. Advances in SELEX and application of aptamers in the central nervous system. Biomol Eng. 2007;24:583–592. doi: 10.1016/j.bioeng.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Mairal T, Ozalp VC, Lozano Sanchez P, Mir M, Katakis I, O'Sullivan CK. Aptamers: molecular tools for analytical applications. Anal Bioanal Chem. 2008;390:989–1007. doi: 10.1007/s00216-007-1346-4. [DOI] [PubMed] [Google Scholar]
- 10.Weiss S, Proske D, Neumann M, Groschup MH, Kretzschmar HA, Famulok M, Winnacker EL. RNA aptamers specifically interact with the prion protein PrP. J Virol. 1997;71:8790–8797. doi: 10.1128/jvi.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proske D, Gilch S, Wopfner F, Schatzl HM, Winnacker EL, Famulok M. Prion-protein-specific aptamer reduces PrPSc formation. Chembiochem. 2002;3:717–725. doi: 10.1002/1439-7633(20020802)3:8<717::AID-CBIC717>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 12.Rhie A, Kirby L, Sayer N, Wellesley R, Disterer P, Sylvester I, Gill A, Hope J, James W, Tahiri-Alaoui A. Characterization of 2′-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J Biol Chem. 2003;278:39697–39705. doi: 10.1074/jbc.M305297200. [DOI] [PubMed] [Google Scholar]
- 13.Sayer NM, Cubin M, Rhie A, Bullock M, Tahiri-Alaoui A, James W. Structural determinants of conformationally selective, prion-binding aptamers. J Biol Chem. 2004;279:13102–13109. doi: 10.1074/jbc.M310928200. [DOI] [PubMed] [Google Scholar]
- 14.Sekiya S, Nishikawa F, Noda K, Kumar PK, Yokoyama T, Nishikawa S. In vitro selection of RNA aptamers against cellular and abnormal isoform of mouse prion protein. Nucleic Acids Symp Ser (Oxf) 2005:361–362. doi: 10.1093/nass/49.1.361. [DOI] [PubMed] [Google Scholar]
- 15.Mercey R, Lantier I, Maurel MC, Grosclaude J, Lantier F, Marc D. Fast, reversible interaction of prion protein with RNA aptamers containing specific sequence patterns. Arch Virol. 2006;151:2197–2214. doi: 10.1007/s00705-006-0790-3. [DOI] [PubMed] [Google Scholar]
- 16.Sekiya S, Noda K, Nishikawa F, Yokoyama T, Kumar PK, Nishikawa S. Characterization and application of a novel RNA aptamer against the mouse prion protein. J Biochem. 2006;139:383–390. doi: 10.1093/jb/mvj046. [DOI] [PubMed] [Google Scholar]
- 17.Takemura K, Wang P, Vorberg I, Surewicz W, Priola SA, Kanthasamy A, Pottathil R, Chen SG, Sreevatsan S. DNA aptamers that bind to PrP(C) and not PrP(Sc) show sequence and structure specificity. Exp Biol Med (Maywood) 2006;231:204–214. doi: 10.1177/153537020623100211. [DOI] [PubMed] [Google Scholar]
- 18.Nishikawa F, Murakami K, Noda K, Yokoyama T, Nishikawa S. Detection of structural changes of RNA aptamer containing GGA repeats under the ionic condition using the microchip electrophoresis. Nucleic Acids Symp Ser (Oxf) 2007:397–398. doi: 10.1093/nass/nrm199. [DOI] [PubMed] [Google Scholar]
- 19.Bibby DF, Gill AC, Kirby L, Farquhar CF, Bruce ME, Garson JA. Application of a novel in vitro selection technique to isolate and characterise high affinity DNA aptamers binding mammalian prion proteins. J Virol Methods. 2008;151:107–115. doi: 10.1016/j.jviromet.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Ogasawara D, Hasegawa H, Kaneko K, Sode K, Ikebukuro K. Screening of DNA Aptamer Against Mouse Prion Protein by Competitive Selection. Prion. 2008;1:248–254. doi: 10.4161/pri.1.4.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 22.Bagga PS, Wilusz J. Northwestern screening of expression libraries. Methods Mol Biol. 1999;118:245–256. doi: 10.1385/1-59259-676-2:245. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi H, Takata M, Iwamaru Y, Ushiki Y, Kimura KM, Tagawa Y, Shinagawa M, Yokoyama T. Effect of tissue deterioration on postmortem BSE diagnosis by immunobiochemical detection of an abnormal isoform of prion protein. J Vet Med Sci. 2004;66:515–520. doi: 10.1292/jvms.66.515. [DOI] [PubMed] [Google Scholar]
- 24.Matsugami A, Ouhashi K, Kanagawa M, Liu H, Kanagawa S, Uesugi S, Katahira M. An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J Mol Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 25.Laughlan G, Murchie AI, Norman DG, Moore MH, Moody PC, Lilley DM, Luisi B. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science. 1994;265:520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- 26.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 27.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Catasti P, Chen X, Moyzis RK, Bradbury EM, Gupta G. Structure-function correlations of the insulin-linked polymorphic region. J Mol Biol. 1996;264:534–545. doi: 10.1006/jmbi.1996.0659. [DOI] [PubMed] [Google Scholar]
- 29.Nandi PK. Interaction of prion peptide HuPrP106-126 with nucleic acid. Arch Virol. 1997;142:2537–2545. doi: 10.1007/s007050050261. [DOI] [PubMed] [Google Scholar]
- 30.Nandi PK, Nicole JC. Nucleic acid and prion protein interaction produces spherical amyloids which can function in vivo as coats of spongiform encephalopathy agent. J Mol Biol. 2004;344:827–837. doi: 10.1016/j.jmb.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 31.Liu H, Kugimiya A, Matsugami A, Katahira M, Uesugi S. Quadruplex structures of RNA 14-mer, r(GGAGGUUUUGGAGG) and DNA 14-mer, d(GGAGGTTTTGGAGG) Nucleic Acids Res Suppl. 2002;2:177–178. doi: 10.1093/nass/2.1.177. [DOI] [PubMed] [Google Scholar]
- 32.Zeiler B, Adler V, Kryukov V, Grossman A. Concentration and removal of prion proteins from biological solutions. Biotechnol Appl Biochem. 2003;37:173–182. doi: 10.1042/ba20020087. [DOI] [PubMed] [Google Scholar]
- 33.Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 34.Prusiner SB, Groth DF, Bolton DC, Kent SB, Hood LE. Purification and structural studies of a major scrapie prion protein. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- 35.Caughey B, Raymond GJ, Ernst D, Race RE. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prusiner SB, McKinley MP, Groth DF, Bowman KA, Mock NI, Cochran SP, Masiarz FR. Scrapie agent contains a hydrophobic protein. Proc Natl Acad Sci USA. 1981;78:6675–6679. doi: 10.1073/pnas.78.11.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luhrs T, Zahn R, Wuthrich K. Amyloid formation by recombinant full-length prion proteins in phospholipid bicelle solutions. J Mol Biol. 2006;357:833–841. doi: 10.1016/j.jmb.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 39.Gomes MP, Millen TA, Ferreira PS, Cunha ESNL, Vieira TC, Almeida MS, Silva JL, Cordeiro Y. Prion protein complexed to N2a cellular RNAs through its N-terminal domain forms aggregates and is toxic to murine neuroblastoma cells. J Biol Chem. 2008;283:19616–19625. doi: 10.1074/jbc.M802102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davidowitz E, Eljuga L, Dover K, Tian J, Grossman A. Concentration of prion protein from biological samples to increase the limits of detection by immunoassay. Biotechnol Appl Biochem. 2005;41:247–253. doi: 10.1042/BA20040080. [DOI] [PubMed] [Google Scholar]
- 41.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 42.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USA. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]