Abstract
Glucantransferase Bgl2p is a major conserved cell wall constituent described for a wide range of yeast species. In the baker's yeast Saccharomyces cerevisiae it is the only non-covalently bound cell wall protein that cannot be released from cell walls by sequential SDS and trypsin treatment. It contains seven amyloidogenic determinants. Circular dichroism analysis and fluorescence spectroscopy with thioflavin T indicate the presence of β-sheet structures in Bgl2p isolates. Bgl2p forms fibrils, a process that is enforced in the presence of other cell wall components. Thus the data obtained is the first evidence for amyloid-like properties of yeast cell wall protein—glucantransferase Bgl2p.
Key words: glucantransferase Bgl2p, cell wall amyloid-like protein
Introduction
The yeast cell wall consists of mannoproteins, β-glucans and chitin forming a macromolecular complex.1 Cell wall proteins fulfill essential functions as structural components and enzymes involved in cell wall assembly.2–5 With respect to their biochemical characteristics two groups of yeast cell wall proteins can be discriminated. The first group includes proteins that can be removed from the cell walls by SDS/mercaptoethanol treatment at 100°C and are therefore considered to be disulphide-linked or non-covalently attached to the wall components (SEP—SDS-extractable proteins). The second group consists of proteins which are covalently linked to the glucan framework (CWP—covalently bound cell wall proteins).1,2
Amyloids are widely spread on cell surface of filamentous fungi and bacteria providing a functional coat for these microorganisms6,7 and also were related to aerial growth and dispersion of spores.8 Polypeptides forming such fibrils include unrelated proteins like hydrophobins and chaplins. A potential role has been discussed for such amyloids in context of human and animal infections by pathogenic microorganisms.7 Yeast prions are under thorough investigation.9 So far, amyloid-like proteins have not been described for the cell surface of yeast species. Here we demonstrated that the cell wall-residing glucantransferase Bgl2p from Saccharomyces cerevisiae possesses the properties of protein with amyloid-like nature.
Results
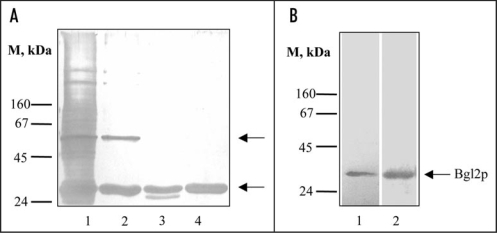
The starting point of this investigation was the computer analysis which allowed us to predict that some yeast cell wall proteins contain potential amyloidogenic determinants. The most pronounced amyloidogenic properties were predicted for two SEPs of S. cerevisiae cell walls: Bgl2p and Exg1p (Table 1). It is well known that S. cerevisiae cell walls contain SEPs that can be extracted into hot Laemmli buffer (Fig. 1A, lane 1). We revealed that a lot of them can be extracted by treatment with 1% SDS at 37°C for an extended period of time, except two proteins which remain in the cell wall after this treatment, and then can be extracted into hot Laemmli buffer (Fig. 1A, lane 2). One of them can be removed by hot water (100°C, 5 minutes) (data not shown). It was immunologically identified as glucantransferase Bgl2p. We demonstrated that Bgl2p is hydrolyzed by proteinase K to a low extent only when attached to the cell wall. In this case Bgl2p can be extracted into hot Laemmli buffer in partly digested form (Fig. 1A, lane 3). Even after digestion under more rigid conditions an obviously proteinase K-resistant core persists (data not shown). Similarly, Bgl2p is poorly hydrolyzed by trypsin despite a high content of lysine and arginine residues (see UniProtKB/Swiss-Prot Entry No. P15703) and can be extracted into hot Laemmli buffer from cell walls treated with trypsin in non-digested form (Fig. 1A, lane 4). We used SDS and trypsin resistance of Bgl2p for its purification (see Materials and methods). Also Bgl2p can be extracted from cell wall with dimethyl sulfoxide (DMSO) used to solubilize amyloids (Fig. 1B), other SEPs haven't been extracted with DMSO from non-treated with trypsin and SDS wild type cell walls (data not shown).
Table 1.
Potential amyloidogenic determinants in S. cerevisiae cell wall proteins
Figure 1.
Analysis of non-covalently bound proteins (SEPs) from the cell wall of S. cerevisiae. (A) SDS-PAGE of SEPs. Lane 1, SEPs extracted from wild type cell wall with hot Laemmli sample buffer (5 min, 100°C). Lane 2, SEPs extracted from cell walls after incubation (1 hour at 37°C) with 1% SDS; lane 3, with 1% SDS and proteinase K (10 mg/ml 1 hour at 37°C) step by step; lane 4, after incubation with 1% SDS and trypsin (2.5 mg/ml 30 min at 37°C) step by step. Upper arrow, presumptive Exg1p (61 kDa); lower arrow, Bgl2p (29 kDa). M stands for molecular weight markers, masses of marker proteins indicated on the figure (kDa). Proteins were separated on two-step 10/12% resolving gel and visualized by silver staining. (B) Silver stained SDS-PAGE (lane 1) and Western blot analysis (lane 2) of DMSO extracts of wild type cell walls. Proteins were extracted from cell walls, separated on 10% SDS gels and transferred to a nitrocellulose membrane. The transferred proteins were probed with an anti-Bgl2p antibody.
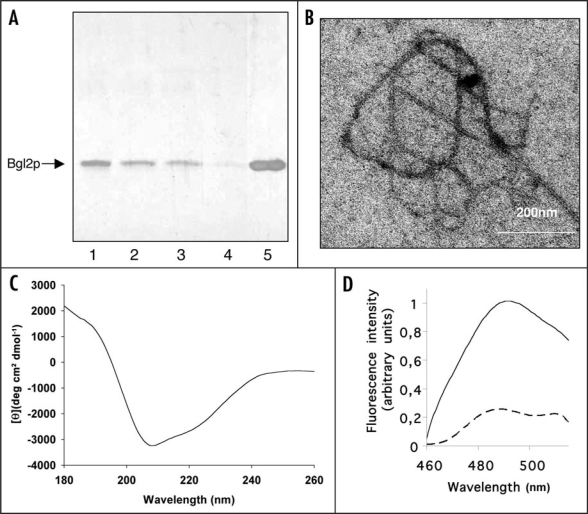
We observed that the share of isolated Bgl2p monomers decreased during incubation at room temperature (Fig. 2A lanes 1–4) if the samples were not boiled after addition of sample buffer and that boiling in sample buffer restored the prevalence of monomers (Fig. 2A lane 5). Electron microscopy inspection of Bgl2p solution after an incubation for 96 h revealed fibrils of about 12 nm width and more than 1 µm length (Fig. 2B). For control we executed parallel experiments using cell wall preparations of the Δbgl2 strain, no fibril-like structures was observed in such control experiments. Based on these findings we suppose that the decrease of Bgl2p monomers most likely corresponds to the formation of high molecular weight structures that cannot enter the gel under the conditions applied to the analysis. Circular dichroism analysis indicated that isolated Bgl2p was rich in β-sheet secondary structure (Fig. 2C; Table 2). Unfortunately we couldn't obtain the CD-spectrum of denaturated Bgl2p since it turns insoluble when heated at 100°C for 5 minutes, in spite of the fact that Bgl2p preserves its β-structural conformation during heating up to 90°C (see Table 2). Furthermore, in fluorescent spectroscopy, an increase of fluorescence at 490 nm was observed in Bgl2p samples treated with thioflavin T (ThT) (Fig. 2D, solid line). Boiling resulted in the disappearance of the signal (Fig. 2D, dashed line).
Figure 2.
Amyloid-like properties of Bgl2p. (A) SDS-PAGE analysis Bgl2p. Bgl2p was extracted from cell wall preparations as described in Materials and Methods. Proteins were visualized by silver staining. After extraction Bgl2p samples were incubated at room temperature for 0 (lane 1), 1 hour (lane 2), 24 hours (lane 3) and 96 hours (lane 4) and applied to SDS-PAGE without boiling after addition of Laemmli buffer. In lane 5 the Bgl2p sample identical to that applied to lane 4 was boiled after buffer addition (5 min, 100°C). (B) Electron microscopy analysis of Bgl2p samples. Bgl2p sample were incubated at room temperature for 96 hours and inspected by electron microscopy. Bgl2p is present as high molecular weight structures forming fibrils (for control we executed parallel experiments using cell wall preparations of the Δbgl2 strain, no fibril-like structures were observed in such control experiments). Negative-staining EM was used. (C) Circular dichroism spectrum of Bgl2p at 20°C. Bgl2p was extracted from cell wall as described in Materials and Methods. The spectrum revealed the high β-sheet structure content in the sample. (D) Fluorescence emission spectra of Bgl2p-bound thioflavin T. Bgl2p was extracted from cell wall preparations (solid line) and the identical sample was boiled for 5 minutes before applying (dashed line). Autofluorescence of protein in the absence of ThT was negligible. Excitation wavelength is 435 nm.
Table 2.
The results of analysis of Bgl2p circular dichroism spectra for secondary structure using DICHRO WEB online server
| Algorithm and conditions used | Share of secondary structure elementsa | ||||||
| αR | αD | βR | βD | T | U | ||
| CDSSTR35 | 20°C | −0,02 | 0,03 | 0,30 | 0,12 | 0,21 | 0,34 |
| SELCON336 | 20°C | 0,049 | 0,191 | 0,275 | 0,149 | 0,065 | 0,260 |
| 90°C | 0,003 | 0,060 | 0,228 | 0,104 | 0,161 | 0,280 | |
| CONTINLL37 | 20°C | 0,013b | 0,064 | 0,251 | 0,119 | 0,238 | 0,314 |
| 90°C | 0,039 | 0,074 | 0,239 | 0,116 | 0,223 | 0,309 | |
| 20°C | 0,004c | 0,046 | 0,276 | 0,137 | 0,219 | 0,319 | |
| 90°C | 0,005 | 0,048 | 0,290 | 0,141 | 0,210 | 0,307 | |
αR, regular α-helix; αD, distorted α-helix; βR, regular β-strand; βD, distorted β-strand, T, turn; U, unordered.
Closest matching solution with all proteins of reference set.
Average of all matching solutions.
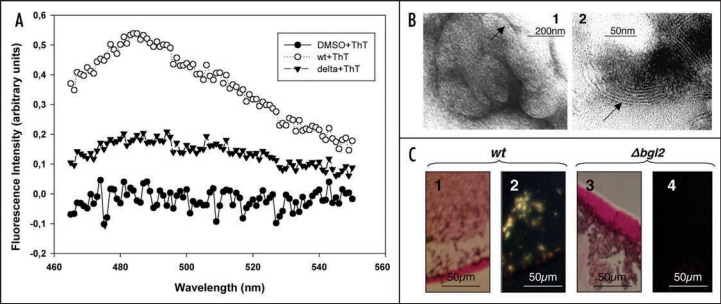
To minimize the probability that the isolation procedure gave rise to Bgl2p fibril formation and to obtain the additional evidence of amyloid nature of Bgl2p we analyzed ThT fluorescence of DMSO extracts obtained from wild type and Δbgl2 mutant strain cell walls.
The data obtained demonstrated that extract from wild type cell walls gave the fluorescence at 485 nm (Fig. 3A). In contrast DMSO-extract from Δbgl2 cell walls gave no peak. Bgl2p amyloid fibrils are formed more efficiently when the protein is extracted from cell walls and then incubated in the presence of cell wall preparations. We demonstrated that 6 nm wide fibrillar structures of finger print resemblance could be detected by electron microscopy after incubation of purified Bgl2p with cell wall isolates that were partially digested with glucanase (Fig. 3B). The preparation containing extracts obtained from cell walls of the Δbgl2 mutant strain lacked such fibrils. When Bgl2p is extracted from cell walls and then incubated in the presence of cell wall preparations it specifically binds the amyloidophilic dye Congo Red resulting in green birefringence of polarized light (Fig. 3C).
Figure 3.
Analysis of amyloid-like proteins in cell wall preparations derived from wild type and Δbgl2 strains. (A) Fluorescence emission spectra of ThT in presence of DMSO extracts obtained from S. cerevisiae wild type (wt + ThT) and Δbgl2 (delta + ThT) cell walls and DMSO (DMSO + ThT). In all samples DMSO concentration was 2.8%. Autofluorescence of protein in absence of ThT was negligible. Excitation wavelength is 450 nm. (B) Electron microscopy analysis of Bgl2p incubated with cell walls partially digested with glucanase (1 mg/ml 30 min at 37°C). The arrows indicate ‘fingerprint’-like fibrillar structures. Lane 1, a group of ‘fingerprints’; lane 2, individual ‘fingerprint’. Negative-staining EM was used. (C) Light microscopy analysis of extracts obtained from wild type and Δbgl2 cell walls incubated at room temperature for a week in the presence of corresponding cell walls partially digested with glucanase. Congo Red tinted green birefringence in polarized light. Bgl2p solution (1) and control sample obtained from Δbgl2 strain cell walls (3). Polarized light microscopy analysis of the same extracts of the wild type cell walls (2) and Δbgl2 strain cell walls (4).
Discussion
Bgl2p is a major cell wall protein of different yeast species. In pathogenic yeasts it participates in virulence10 and a high Bgl2p antibody titre is a marker of systemic candidiasis.11 Here we demonstrated that Bgl2p possess amyloidogenic features. Firstly—low solubility in SDS and resistance to protease digestion are considered to be indicative of amyloidogenic proteins.12 Bgl2p is rich in β-sheet secondary structure (Fig. 2C; Table 2), a distinctive feature of amyloids,12–14 and Bgl2p samples treated with ThT give an increase of fluorescence which is characteristic for amyloid fibril-bound ThT.
We suppose that Bgl2p is not the only amyloid-like protein in the yeast cell wall. The second SEP which cannot be removed from cell wall isolates (Fig. 1A, lanes 1 and 2) is most likely glucan 1,3-β-glucosidase Exg1p by molecular weight. And we revealed that Exg1p contains eleven, Bgl2p six short amino-acid stretches of potential amyloidogenic determinants. Such stretches were also found in some additional cell wall proteins, but were of insufficient number and length to define the proteins as amyloidogenic (data not shown). But we restricted our study to Bgl2p, the protein of a more pronounced protease resistance.
We suppose that fibril formation is not restricted to purified isolates but that Bgl2p proteins reside in the cell wall as fibrillar structures however this suggestion needs further experimental confirmation.
It is not unlikely that interaction with other cell wall components is required for fibril formation. Previously we have demonstrated that Bgl2p molecules can be detected in the growth medium when GPI anchor formation is impaired and Bgl2p presents in yeast cell wall at low level.15 Under such conditions Bgl2p is secreted to the medium in non-fibrillar form (data not shown). This suggests a possible involvement of GPI proteins in the conversion of Bgl2p into amyloid fibrils.
The cell surface amyloids of microorganisms usually perform an important role in cell adaptation to the environment, for example SC3 hydrophobin lowers surface tension at water-air interface allowing filamentous fungi to escape the aqueous phase to form aerial structures.16 Although Bgl2p has been studied since 1989, its physiological role remains obscure. Strain overexpressing BGL2 exhibit a decreased growth rate,17 Δbgl2 mutant strains are characterized by increased chitin levels in the cell wall.18 Deletion of BGL2 gene is not lethal. Moreover viability of Δbgl2 mutant strains is not impaired under standard cultivation conditions and a decreased growth rate of Δscw4 and Δscw10 strains is suppressed when deleting BGL2.19 Similar to the functions of amyloids in other microorganisms Bgl2p may contribute to forming of cell wall assembly, especially under extreme conditions.
Bgl2p from Candida utilis and C. albicans have a similar share of amyloidogenic determinants (0.15 for S. cerevisiae, 0.30 for C. utilis, 0.11 for C. albicans). Bgl2p from C. utilis exhibits properties similar to that of S. cerevisiae. Bgl2p from Hansenula polymorpha as well as that from S. cerevisiae couldn't be extracted from cell wall by incubation with SDS at 37°C (Vitaly V. Kushnirov personal communication, Institute of Experimental Cardiology). Our data provide the first evidence for a protein with amyloidogenic properties in the cell wall of yeasts. Bgl2p is a conserved protein with homologous counterparts described for a wide range of yeast species.17,20,21
Materials and Methods
Prediction of amyloidogenic regions in protein sequence.
For prediction of amyloidogenic regions, we used the previously described method based on large number of contacts per residue in protein sequences.22–24 For each amino acid residue within a protein sequence, an expected number of contacts per residue is defined for the various residue types (which is the average number of contacts at a distance below 8 Å for the given type of residue in 3D structures of proteins).22,23 Then, the values are averaged with a sliding window of seven residues.23 Sequences composed of residues above a threshold value (which is 21,4 expected contacts per residue) are predicted as amyloidogenic if the size of such a region is not smaller than the sliding window.22–24 Thus, the predicted amyloidogenic regions are regions which have a large number of expected contacts per residue.
Yeast strains and growth conditions.
The S. cerevisiae parent strains MAY591,25 (MATα leu2-3,112 lys2-801 ura3-52 his3-Δ200) and DBY 746 (MATα ura3-52 leu2-3,112 trp1-289 his3-Δ1) were used; they are referred to as wild type (or wt) in the text. The mutant strains were obtained in our laboratory by disruption of chromosomal copy of BGL2 gene by insertion of URA3 (MATα leu2-3,112 lys2-801 ura3-52 his3-Δ200 BGL2::URA3 and MATα ura3-52 leu2-3,112 trp1-289 his3-Δ1BGL2::URA3 respectively); they are denoted to as Δbgl2.
The C. utilis strain VKM Y-74 was obtained form All-Russian Collection of Microorganisms—VKM (Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, Pushchino, Moscow Region, Russia).
Yeast cells were grown at 30°C in YPD medium (1% yeast extract, 2% peptone, 2% glucose).
Cell wall isolation.
Cell walls were obtained as previously described.4 Cell disruption was performed with glass beads in presence of 5 mM PMSF and 5 mM EDTA. Cell walls were precipitated by centrifugation at 5000 g and then were washed sequentially with 1% sucrose, 1 M NaCl, 1% NaCl and deionized water.
Purification of Bgl2p.
Bgl2p was purified from cell walls which were incubated with trypsin (1 mg/ml in 50 mM Tris/HCl pH 7.5) and with 1% SDS for 1 hour (37°C). This procedure allows removing all non-covalently attached proteins except Bgl2p from cell walls. A pellet obtained by centrifugation at 5000 g was extracted three times with cold butanol-water (0.7:1, vol:vol) for 15 min (1 volume of cell debris per three volumes of butanol-water). After each extraction the suspension was centrifuged at 10,000 g, the supernatant was discarded. The pellet obtained after the last extraction was washed twice in water. After subsequent washing with 1 M NaCl (four times), 1% NaCl (four times), 1% sucrose (twice) and water cell walls were centrifuged and then suspended in water containing 0.05% NaN3 and 5 mM PMSF for storage. The pellet was re-suspended in water and incubated at 55°C for 15 min, centrifuged and incubated again at 70°C for 15 min and centrifuged at 10,000 g. The final extract contained the pure Bgl2p (25 µg/ml). Control extracts obtained from Δbgl2 mutant strain cell wall preparations with this technique contained no Bgl2p.
Protein concentration determination.
Peptide bond absorption at 205 nm was measured using Varian Cary 300 Bio UV-Visible Spectrophotometer (Varian Inc., USA). Protein concentration was determined according to Scopes.26
Electrophoresis.
Electrophoresis was performed according to Laemmli27 in 10% or two-step 10–12% resolving polyacrylamide gels. Proteins were visualized by silver staining or immunologically detected by Western Blot analysis.
Antibodies against Bgl2p.
Bgl2p antiserum was raised in male BALB/c mice (SPF status) using SDS PAGE-purified protein (40 µg per mouse).28
Thioflavin T fluorescence.
ThT binding assay was performed according to LeVine.29 Fluorescence measurements were carried out with a Cary Eclipse fluorescence spectrophotometer (Varian Inc., USA).
Circular dichroism analysis.
Circular dichroism spectra were obtained using Chirascan Circular Dichroism Spectrometer (Applied Photophysics Ltd., UK) at 20–90°C. A cell with a 0.2 cm pathlength was used for spectra recorded between 180 and 260 nm, with sampling points every 0.5 nm. Five scans were recorded, and baseline spectra were subtracted from each spectrum. Data were processed using Applied Photophysics Chirascan Viewer and SigmaPlot package.12 Analysis of protein circular dichroism spectra for secondary structure was carried out using DICHROWEB online server available at www.cryst.bbk.ac.uk/cdweb/html/dw2.html.30,31 Protein Reference Set 3 (optimized for 185–240 nm) was used.
Congo red binding assay.
Samples were applied to object slides that were thoroughly washed with alcohol and dried at room temperature. Specimens were stained with 0.0002% Congo Red solution and visualized using an Opton microscope in polarized light.12,32
Electron microscopy (EM).
Negative-staining EM was used. 2 µl samples were absorbed onto glow-discharged carbon-coated, Formvar-filmed 400-mesh copper grids and immediately dried down. 2% uranyl acetate staining solution was then absorbed for 2 min. Grids were allowed to dry in a light-protected environment and viewed in a JEM-100B (JEOL, Japan) electron microscope at the accelerating potential of 80 kV.33
Other methods.
Yeast cell walls were partially digested with glucanase (laminarinase, Sigma, 1 mg/ml 30 min at 37°C) and with proteinase K (Serva, 10 mg/ml 1 hour at 37°C). Prior to use in further experiments the enzymes were washed out.
DMSO extraction of Bgl2p from yeast cell walls was performed as described earlier for dissolution of amyloid fibrils with some modifications.34
Acknowledgements
We thank S.A. Kuznetsov (University of Rostock, Rostock, Germany) for his support in frame of an agreement between Lomonosov Moscow State University and the University of Rostock. Also we thank V.V. Shubin (Bach Institute of Biochemistry, Russian Academy of Sciences, Moscow, Russia) for the invaluable help in obtaining and interpreting the circular dichroism spectra and O.S. Morenkov (Institute of Cell Biophysics, Russian Academy of Sciences, Pushchino, Russia) for providing antibodies against Bgl2p. This work was funded by the grants 06-04-49262 and 08-04-00561 from Russian Foundation for Basic Research, by Russian Federation President's grant for government support of leading scientific schools, by the Russian Academy of Sciences (“Molecular and Cell Biology” program), by the “Russian Science Support Foundation” and by the INTAS grants 01-0583 and 05-1000004-7747.
Abbreviations
- Δbgl2
mutant strain without Bgl2p
- Bgl2p
glucantransferase
- CD
circular dichroism
- CWP
covalently bound cell wall proteins
- DMSO
dimethyl sulfoxide
- EDTA
disodium ethylenediaminetetraacetate
- EM
electron microscopy
- Exg1p
glucan 1,3-β-glucosidase
- kDa
kilodalton
- NaCl
sodium chloride
- NaN3
sodium azide
- OD
optical density
- PAGE
polyacrylamide gel electrophoresis
- PMSF
phenylmethylsulphonyl fluoride
- SDS
sodium dodecyl sulfate
- SEP
SDS-extractable proteins
- ThT
thioflavin T
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/6645
References
- 1.Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapteyn JC, Van Den Ende H, Klis FM. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim Biophys Acta. 1999;1426:373–383. doi: 10.1016/s0304-4165(98)00137-8. [DOI] [PubMed] [Google Scholar]
- 3.Mrša V, Tanner W. Role of NaOH-extractable cell wall proteins Ccw5p, Ccw6p, Ccw7p and Ccw8p (members of the Pir protein family) in stability of the Saccharomyces cerevisiae cell wall. Yeast. 1999;15:813–820. doi: 10.1002/(SICI)1097-0061(199907)15:10A<813::AID-YEA421>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Klebl F, Tanner W. Molecular cloning of a cell wall exo-beta-1,3-glucanase from Saccharomyces cerevisiae. J Bacteriol. 1989;171:6259–6264. doi: 10.1128/jb.171.11.6259-6264.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popolo L, Vai M. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim Biophys Acta. 1999;1426:385–400. doi: 10.1016/s0304-4165(98)00138-x. [DOI] [PubMed] [Google Scholar]
- 6.Wösten HA, de Vocht ML. Hydrophobins, the fungal coat unravelled. Biochim Biophys Acta. 2000;1469:79–86. doi: 10.1016/s0304-4157(00)00002-2. [DOI] [PubMed] [Google Scholar]
- 7.Gebbink MF, Claessen D, Bouma B, Dijkhuizen L, Wösten HA. Amyloids— a functional coat for microorganisms. Nature Rev Microbiol. 2005;3:333–341. doi: 10.1038/nrmicro1127. [DOI] [PubMed] [Google Scholar]
- 8.Wösten HA. Hydrophobins: multipurpose proteins. Annu Rev Microbiol. 2001;55:625–646. doi: 10.1146/annurev.micro.55.1.625. [DOI] [PubMed] [Google Scholar]
- 9.Ter-Avanesyan M, Derkatch I, Baskakov I, Kushnirov V. Unraveling prion structures and biological functions. Genome Biol. 2005;6:366. doi: 10.1186/gb-2005-6-13-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarthy AV, McGonigal T, Coen M, Frost DJ, Meulbroek JA, Goldman RC. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-beta-glucosyltransferase. Microbiology. 1997;143:367–376. doi: 10.1099/00221287-143-2-367. [DOI] [PubMed] [Google Scholar]
- 11.Pitarch A, Jiménez A, Nombela C, Gil C. Decoding serological response to Candida cell wall immunome into novel diagnostic, prognostic, and therapeutic candidates for systemic candidiasis by proteomic and bioinformatic analyses. Mol Cell Proteomics. 2006;5:79–96. doi: 10.1074/mcp.M500243-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claessen D, Rink R, de Jong W, Siebring J, de Vreugd P, Boersma FG, Dijkhuizen L, Wosten HA. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003;17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalebina TS, Laurinavichiute DK, Packeiser AN, Morenkov OS, Ter-Avanesyan MD, Kulaev IS. Correct GPI-anchor synthesis is required for the incorporation of endoglucanase/glucanosyltransferase Bgl2p into the Saccharomyces cerevisiae cell wall. FEMS Microbiol Lett. 2002;210:81–85. doi: 10.1111/j.1574-6968.2002.tb11163.x. [DOI] [PubMed] [Google Scholar]
- 16.Wösten HA, van Wetter MA, Lugones LG, van der Mei HC, Busscher HJ, Wessels JG. How a fungus escapes the water to grow into the air. Curr Biol. 1999;9:85–88. doi: 10.1016/s0960-9822(99)80019-0. [DOI] [PubMed] [Google Scholar]
- 17.Mrša V, Klebl F, Tanner W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase. J Bacteriol. 1993;175:2102–2106. doi: 10.1128/jb.175.7.2102-2106.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalebina TS, Farkaš V, Laurinavichiute DK, Gorlovoy PM, Fominov GV, Bartek P, Kulaev IS. Deletion of BGL2 results in an increased chitin level in the cell wall of Saccharomyces cerevisiae. Antonie Leeuwenhoek. 2003;84:179–184. doi: 10.1023/a:1026034123673. [DOI] [PubMed] [Google Scholar]
- 19.Cappellaro C, Mrša V, Tanner W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J Bacteriol. 1998;180:5030–5037. doi: 10.1128/jb.180.19.5030-5037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaffin WL, Lopez-Ribot JL, Casanova M, Gozalbo D, Martinez JP. Cell wall and secreted proteins of Candida albicans: identification, function and expression. Microbiol Mol Biol Rev. 1998;62:130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouyna I, Hartland RP, Fontaine T, Diaquin M, Simenel C, Delepierre M, Henrissat B, Latgé JP. A 1,3-beta-glucanosyltransferase isolated from the cell wall of Aspergillus fumigatus is a homologue of the yeast Bgl2p. Microbiology. 1998;144:3171–3180. [Google Scholar]
- 22.Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. A search for amyloidogenic regions in protein chains. Mol Biol (Moscow) 2006;40:821–828. [PubMed] [Google Scholar]
- 23.Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. Is it possible to predict amyloidogenic regions from sequence alone? Journal of Bioinformatics and Computational Biology. 2006;4:373–388. doi: 10.1142/s0219720006002004. [DOI] [PubMed] [Google Scholar]
- 24.Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. Prediction of amyloidogenic and disordered regions in protein chains. PLoS Computational Biology. 2006;2:177. doi: 10.1371/journal.pcbi.0020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- 26.Scopes RK. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974;59:277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Morenkov OS, Mantsyghin Yu A, Sergeev VA, Sobko Yu A, Morenkova MA, Panchenko OA. The isolation and characteristics of monoclonal antibodies to the glycoprotein GII of Aujeszky's disease virus and their use for the epitopic mapping of GII. Vopr Virusol (Russ) 1994;4:174–177. [PubMed] [Google Scholar]
- 29.LeVine H. Thioflavine T interaction with synthetic Alzheimer's disease {beta}-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 31.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analysis from circular dichroism spectroscopic data. Nucleic Acids Research. 2004;32:668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balbirnie M, Grothe R, Eisenberg DS. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated β-sheet structure for amyloid. Proc Natl Acad Sci USA. 2001;98:2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchard BJ, Chen A, Rozeboom LM, Stafford KA, Weigele P, Ingram VM. Efficient reversal of Alzheimer's disease fibril formation and elimination of neurotoxicity by a small molecule. Proc Natl Acad Sci U S A. 2004;101:14326–14332. doi: 10.1073/pnas.0405941101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirota-Nakaoka N, Hasegawa K, Naiki H, Goto Y. Dissolution of beta2-microglobulin amyloid fibrils by dimethylsulfoxide. J Biochem. 2003;134:159–164. doi: 10.1093/jb/mvg124. [DOI] [PubMed] [Google Scholar]
- 35.Manavalan P, Johnson WC., Jr Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal Biochem. 1987;167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 36.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 37.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]