Abstract
Amyloid refers to the abnormal fibrous, extracellular, proteinaceous deposits found in organs and tissues. Amyloid is insoluble and is structurally dominated by β-sheet structure. Unlike other fibrous proteins it does not commonly have a structural, supportive or motility role but is associated with the pathology seen in a range of diseases known as the amyloidoses. These diseases include Alzheimer's, the spongiform encephalopathies and type II diabetes, all of which are progressive disorders with associated high morbidity and mortality. Not surprisingly, research into the physicochemical properties of amyloid and its formation is currently intensely pursued. In this chapter we will highlight the key scientific findings and discuss how the stability of amyloid fibrils impacts on bionanotechnology.
Key words: fibril, β-sheet, aggregation, Alzheimer's disease, fibre diffraction, ssNMR, small angle X-ray scattering
Introduction
Amyloid fibrils are formed by normally soluble proteins, which assemble to form insoluble fibers that are resistant to degradation. Their formation can accompany disease and each disease is characterized by a specfic protein or peptide that aggregates. Well known examples of amyloid diseases include Alzheimer's disease, Diabetes type 2 and the spongiform encephalopathies (e.g., Mad cow disease). The amyloid fibrils are deposited extracellularly in the tissues and are thought to have a pathogenic effect.1 The fibrillar assemblies are inherently stable and structural studies have revealed that they are composed predominantly of β-sheet structure in a characteristic cross-β conformation. Recently, a number of examples of functional amyloid have been identified including a constituent of melanosomes, curli and hydrophobins.2
The History of Amyloid
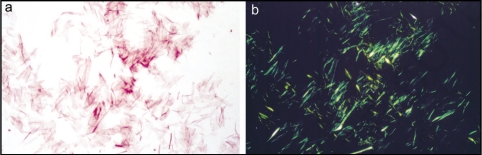
The term ‘amyloid’ was coined initially by Schleiden and then by Virchow in the mid-19th century to describe the iodine stained deposits seen in liver at autopsy. Consequently the deposits were thought to be carbohydrate in nature until their high nitrogen content was later established.3 Nevertheless, the inaccurately descriptive name was retained for these highly proteinaceous deposits. Further tinctorial properties included the specific binding of amyloid to the dye Congo Red which produced an apple green birefringence when examined between cross polarisers in a light microscope4 (Fig. 1). This finding suggested that amyloid was fibrillar in structure and subsequent transmission electron micrographs of amyloid confirmed this.5 Further progress in biochemical and biophysical techniques enabled the isolation of amyloid fibrils from tissues6 and their now characteristic cross-β structure was interpreted from X-ray fiber diffraction patterns.7
Figure 1.
Isolated amyloid fibrils composed of Aα chain fragment of fibrinogen (a) stained with Congo red and visualized by light microscopy and (b) between crossed polars, showing characteristic apple-green birefringence. Figure adapted from reference 83.
Amino acid composition and sequence analysis of the proteins comprising a range of ex vivo amyloid fibrils revealed that each amyloid disorder was associated with a particular protein or peptide.8 To date, more than 20 plasma proteins have been identified that form amyloid (Table 1). Interestingly, despite the obvious differences in amino acid sequences and native structure, these amyloidogenic peptides all appear to share a common β-sheet conformation of their polypeptide backbone7,9 and it is likely that this characteristic confers the fibrillar, proteolytic resistant and insoluble characteristics to all forms of amyloid.
Table 1.
Some amyloidoses and their respective precursors and amyloid-ogenic proteins
| Disease | Precursor protein | Amyloid protein |
| Alzheimer's disease | Amyloid precursor protein | Aβ peptides |
| Atrial amyloidosis | Atrial natriuretic factor (ANF) | Amyloid ANF |
| Spongiform encephalopathies | Prion protein (PrPc) | PrPsc |
| Primary systemic amyloidosis | Immunoglobulin light and heavy chains | AL and AH |
| Senile systemic amyloidosis | Wild-type transthyretin | ATTR |
| Haemodialysis-related amyloidosis | β2-microglobulin | Aβ2M |
| Hereditary nonneuropathic systemic amyloidosis | Lysozyme | ALys |
| Type II diabetes | Pro-IAPP | IAPP or “amylin” |
| Injection-localized amyloidosis | Insulin | AIns |
| Secondary systemic amyloidosis | (Apo) serum amyloid A | Serum amyloid A |
| Hereditary cerebral amyloid angiopathy | Cystatin C | ACys |
| Finnish hereditary systemic amyloidosis | Gelsolin | AGel |
| Familial amyloid polyneuropathy I | Transthyretin variants | ATTR |
| Familial amyloid polyneuropathy II | Apolipoprotein A1 | AApoA1 |
| Ageing pituitary, prolactinomas | Prolactin | APro |
| Familial amyloidosis | Fibrinogen αA-chain | AFib |
| British familial dementia | Amyloid Bri Precursor Protein | ABri |
The finding that lysosomal extracts digested immunoglobulin light chain precursor protein into amyloidogenic fragments10 led to the realisation that many amyloid forming peptides were produced by the proteolytic processing of a precursor protein (Table 1). By 1982, Prusiner put forward the ‘prion’ hypothesis that described an infectious protein particle capable of causing scrapie (a fatal neurodegenerative disease) in sheep.11 This protein was amyloidogenic but unlike other amyloid forming proteins it was infective and was termed: prion. Two years later, Aβ, the peptide forming the bulk of amyloid plaques in Alzheimer's diseased brain specimens, was identified and biochemically characterized.12 In 1986, attention was refocused on prions when a devastating epidemic affected cattle in the UK. These animals were dying of Bovine spongiform encephalopathy (BSE), a prion disease with links to scrapie prion from sheep. Ten years later, a new fatal, neurodegenerative prion disease emerged in humans, variant Creutzfeldt-Jacob disease (vCJD) and the transmissibility of prions from species to species could not be denied as vCJD was strongly linked with exposure to the BSE agent. Interest in prion diseases and amyloid formation (believed to be important to the pathogenesis of the diseases) now escalated.
Earlier biochemical analysis of amyloid deposits had demonstrated that amyloid was not merely composed of fibrils but also contained the serum amyloid protein (SAP) and proteoglycans. Although SAP is believed to have a scavenging function whereby it may stabilize amyloid formation,13 the role of these proteins in amyloid formation and deposition is yet to be discerned. In 1988, the Pepys laboratory radiolabelled SAP and used it as an in vivo tracer to follow the progress of amyloid disorders in patients.14
Three years later, the propensity of a peptide to form amyloid was found to be sequence dependent. Polypeptides containing glutamine repeats were found to be associated with some inherited, neurodegenerative amyloidoses.15 Subsequently, Kelly and coworkers found that purified transthyretin formed fibrils in vitro when its native state was partially unfolded under acidic conditions.16 This finding suggested that induced conformational changes were sufficient to trigger aggregation into amyloid fibrils and implied that erroneous protein folding or misfolding underpinned the mechanism of amyloid formation, fuelling a new area of intense research in protein chemistry.
Amyloidogenesis: Assembly of Amyloidogenic Proteins
With advances in solid phase protein synthesis techniques in the early nineties it was possible to use synthetic Alzheimer's related peptide, Aβ(1–42), its truncated homologues and other short amyloid forming peptides to produce amyloid-like fibrils.17 These were physically, morphologically and tinctorially similar to ex vivo amyloid fibrils and enabled the study of amyloid ultrastructure and formation. An extensive range of biophysical tools and techniques including nuclear magnetic resonance (NMR), Circular Dichroism (CD), X-ray fiber diffraction, atomic force and electron microscopy (AFM and EM), Fourier Transform Infrared Spectroscopy (FTIR) have all since contributed to a better understanding of the structure of amyloid fibrils at a molecular level.
Studying the mechanism of formation of amyloid fibrils is of major importance since insights into the mechanisms underlying polymerisation of soluble, monomeric peptide into mature insoluble fibrils may provide researchers with possible therapeutic approaches to halting, reversing or avoiding fibril formation. From milestone studies, it is known that conversion of soluble peptide to insoluble amyloid often involves the production of a partially unfolded intermediate.18–20 This thermodynamically unfavorable state rapidly progresses to the stable amyloidogenic form and the kinetics of this transition have been studied in vitro using a range of biophysical methods including light scattering, size exclusion chromatography, fluorimetry and ultracentrifugation. The results of such studies propose a nucleation dependent polymerisation model to describe fibril formation, likening the process to crystallization.21 A heterogenous nucleus (‘seed') or peptide micelle form above a critical ‘threshold’ concentration and fibrils nucleate within these, elongating by irreversibly binding monomers to their free ends.22,23 Fibril growth may be represented diagrammatically as a lag exponential growth curve where the phase is considerably shortened in the presence of seeds.
In the evolution of mature fibrils, several metastable intermediates have been identified and isolated.22,24–26 These include very early species: dimers, trimers, tetramers (collectively known as oligomers27), Aβ derived diffusible ligands or ADDLs28 and the later bead-like structures up to 200 nm in length called protofibrils.29 Alternatively, fibril formation may follow an ‘offset pathway’ without the production of fibrils but instead involving conversion of the intermediates into amorphous deposits.
The dynamics of protein folding/misfolding appear to play a key role in in vitro fibril formation and consequently several molecular mechanisms to explain amyloid formation have been proposed; including the polar zipper30 and domain swapping models.31,32 It has been proposed that amyloid formation is a generic property of all peptides33 since under denaturing conditions many normally globular, nondisease related proteins, have been shown to assemble to form fibrils.34,35 However, the propensity of a peptide to form amyloid is dependent on several factors including: polypeptide charge, sequence, hydrophobicity and secondary structure. Consequently several algorithms have been developed to predict the propensity and the rate at which different sequences will aggregate and which mutations will result in an increase or decrease of aggregation rate.36–40 These algorithms will prove useful in determining which regions of a polypeptide chain is specifically involved in fibril formation or the amyloid core structure.
The Structure of the Amyloid Fibril
Amyloid fibrils are insoluble and heterogeneous, so commonly used methods of structure determination are difficult. Therefore, most studies have involved X-ray fiber diffraction, electron microscopy41 and more recently, solid state nuclear magnetic resonance (ssNMR)42 and electroparamagnetic resonance.43–45
Electron and atomic force microscopy have given insights into the macromolecular structure of amyloid fibrils and have shown that fibrils are long, straight and unbranching (Fig. 2A) and are made up of individual subunits named “protoflaments.”46–48 These may vary in number and are often observed to twist around one another to form the mature fibril.46,48–50 Synthetic amyloid-like fibrils, may vary in morphology and this may depend upon the assembly conditions.46,51
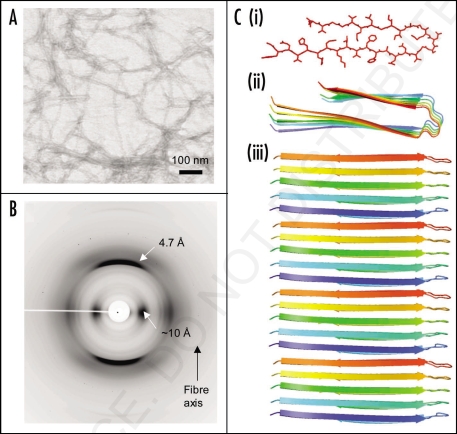
Figure 2.
Synthetic amyloid fibrils made from Aβ peptide (A) electron micrograph showing long, straight, unbranching fibrils. (B) X-ray fiber diffraction pattern from partially aligned amyloid fibrils showing the characteristic “cross-β” diffraction pattern. (C) The structure of the Aβ amyloid fibril interpreted from ssNMR data,67 showing the top view of the fiber (i and ii) with side chains (i), showing the importance of side chain packing with in the fiber and as a cartoon (ii). The side view (iii) revealing the β-strands running perpendicular to the fiber axis.
Cryo-electron microscopy and single particle processing of mature amyloid fibrils composed of the SH3 domain of phosphotidylinositol-3′-kinase indicated that a single fibril was comprised of four protofilaments wound around a central core.2 Consequently, a molecular model of a SH3 amyloid fibril has been generated showing the protofilaments each composed of continuous β-sheet structure.48 Further cryo-electron microscopy studies revealed that synthetic amyloid fibrils formed by insulin,50 lysozyme49 and Aβ(1–42)52 are also composed of several protofilaments wound around one another. The numbers of protofilaments can differ from 2 to 6.50
Early studies on amyloid fibrils revealed that they shared a common X-ray diffraction fingerprint7 and this was later confirmed in a fiber diffraction study of a number of different ex-vivo and synthetic amyloid fibrils.9,53,54 The X-ray diffraction pattern given by amyloid fibrils is “cross-β” (Fig. 2B), a diffraction fingerprint first identified for silk from the egg stalk of the lacewing, Chrysopa.55 The pattern indicates that these fibrous molecules share a particular core structure consisting of β-sheet conformation in which the hydrogen bonding direction runs parallel to the fiber axis and the β-strands are perpendicular, much like the rungs of a ladder (Fig. 2). The diffraction pattern consists of two major reflections at 4.7 Å and 10 Å found on orthogonal axes and arising from the hydrogen bonding distances between β-strands and side chain packing between the sheets respectively (see Fig. 2).9,56 More detailed X-ray diffraction patterns have been obtained from synthetic amyloid fibrils and the additional information obtained has enabled molecular models to be proposed.57 Generally these models are cross-β in nature and the β-sheet conformation forms the core of the structure. The β-strands are hydrogen bonded along the length of the β-sheet structure and parallel to the fiber axis.58 The β-sheet ribbons are associated via side chain interactions57 that serve to stabilize the structure. Very recently, a number of short amyloidogenic peptides have been crystallized this has enabled their atomic structure to be elucidated by X-ray crystallography.59,60 This work has given insights into the nature of the interactions that drive the association of the β-sheets via the amino acid side chains.
SsNMR structural studies have significantly added to our knowledge of the structure of the amyloid fibril61 and this work has been complemented by results from EPR on amyloid formed by a number of different disease associated peptides.45 SsNMR of amyloid fibrils formed by various peptides homologous to regions of the Alzheimer's peptide, Aβ have shown that β-sheet may be arranged parallel or anti-parallel within the protofilaments depending on the properties of the precursor polypeptide.62–66 Studies of fibrils formed from full-length Aβ, have shown that the peptide folds into a β-bend structure that then associates with other molecules to form parallel, in register β-structure67,68 (Fig. 2C).
Small angle X-ray scattering has recently contributed insights into both the structure and the assembly of amyloid fibrils69 revealing for the first time, the oligomeric assembly of insulin fibrils. The SAX study suggested that insulin fibrils are assembled from a helical fibrillation precursor composed of five to six insulin monomers. The mature amyloid fibril is composed of three individual filaments that wrap around one another69 (Fig. 3B). Small angle X-ray scattering from fibrils formed by alpha-synuclein revealed a contrasting structure in which a single filament appears to make up the mature fiber (Vestergaard, personal communication) (Fig. 3A).
Figure 3.
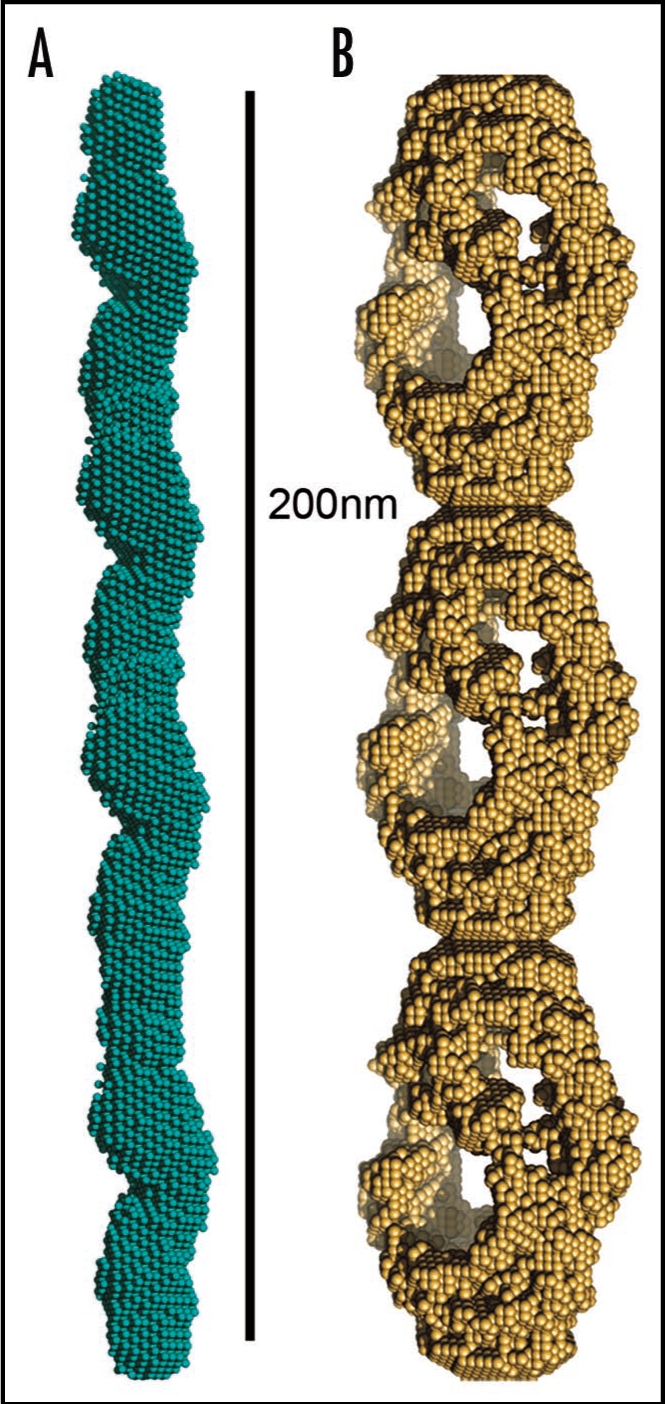
Models of mature protein fibrils based on Small-Angle X-ray scattering solution data. (A) Human alpha-synuclein fibrils and (B) human insulin fibrils.69 The results suggest that insulin fibrils (B) are formed of three intertwining protofibrils, whereas a-synuclein fibril (A) consist of only one protofibril. Each protofibril is assumed to consist of two intertwining protofilaments. Four and three repeating units are shown for alpha-synuclein and insulin respectively.
Toxicity of Amyloid: The Search for the Toxic Species and Its Mode of Action
It is clear that amyloid deposition is a consequence of the amyloidoses; what is uncertain is whether it is a causative agent in its pathogenesis or a secondary event. The amyloid cascade hypothesis70 proposed that altered metabolism of amyloid precursor protein (APP) initiates the pathogenesis of Alzheimer's disease (AD) leading to aggregation of Aβ and formation of neuritic plaques. These plaques would cause further pathological changes including the formation of neurofibrillary tangles and compromised synaptic connections ultimately resulting in neuronal cell loss and dementia. However, there was no correlation between the density of plaques and tangles and the severity of AD. Instead, the concentration of soluble Aβ appeared to correlate with cognitive impairment in other studies.71–73 This finding set the premise for studies suggesting that soluble nonfibrillar intermediates, such as oligomers (20 to >50 kDa globular aggregates, including ADDLs) and protofibrils (curvilinear structures 4–11 nm in diameter and ≤200 nm long) are the actual initiators of AD pathogenesis and that mature fibril formation represents the end point of the disease.
Usually small organic compounds are screened and selected for their binding and inhibitory effects on amyloid precursors or the enzymes involved in their proteolytic processing. These potential anti-amyloid drugs are often highly toxic and have a profound effect on the immune system. Biological ligands such as monoclonal antibodies have been raised against amyloid and are more successful at plaque clearance and reduction in soluble Aβ levels in the CNS.74 However during phase II trials of an Aβ vaccine, 6% of subjects developed an inflammatory response (meningoencephalitis) which halted further clinical development. The search therefore for an effective and safe anti-amyloid drug continues. In the interim, several questions remain unanswered: (1) which intermediate species are responsible for the toxicity of amyloid; (2) what is its atomic structure and the exact mechanism of toxicity and (3) how can this toxicity be safely reversed.
“Exploiting Amyloid”
Functional Amyloid.
Amyloid fibrils are extremely stable and resistant to degradation. They have been described as having a similar tensile strength to steel,75 a property that they share with their structural cousin, silk. Recently, a number of nonpathogenic, functional forms of amyloid have been identified in bacteria, fungi, insects and mammals.2 Curli is a functional fiber found on the surface of bacteria such as E. coli. The fibers share structural similarities to amyloid and formed by a protein called CsgA. Fibrillization is carefully regulated by a protein machine complex.76,77 Many fungi produce amphipathic proteins called hydrophobins.78 These have the ability to assemble into β-sheet rich fibrils at air-water interfaces and these are thought to play a protective role in fungal structures such as spores and fruiting bodies. The yeast prions have been suggested to play a functional role, forming cytoplasmic fibrillar assemblies that may be associated with a method of heritable information transfer.79
In mammals, a protein called Pmel17 is involved in the biosynthesis of melanin. Recently, the Pmel17 protein has been found to form fibrous structures with all the characteristics of amyloid fibrils and these structures are found at the surface of melanosomes. These are thought to be involved in the templating of melanin biosynthesis.80
Bionanotechnological Applications.
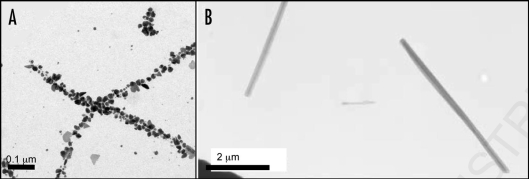
The amyloid fibril has been increasingly examined for its potential role in forming nanotubular scaffolding for bionanotechnology. The fibrils themselves can be very strong, as previously discussed and these fibers can also be functionalized by assembling fusion proteins81 or used as a template for binding to metals.80,82 Baldwin and co-workers81 assembled a fusion protein composed of a functional cytochrome b562 with an amyloidogenic SH3 sequence. The assemblies have the amyloid-like core, displaying functional, folded, globular cytochrome.81 Nanowires have been fabricated by assembling proteins such as the N-terminal region of the yeast prion, Sup35. Conjugate colloidal gold particles were associated along the fibers using exposed cysteine residues of a variant Sup35,80 yielding wires around 100 nm in diameter (Fig. 4A). A very short peptide, composed of two phenylalanine residues, assembles to form amyloid-like nanotubes and these may be functionalised using ionic silver in the centre of the nanotube (Fig. 4B).82 This work yielded nanowires around 20 nm in diameter.
Figure 4.
Amyloid-like fibers for bionanotechnology. (A) Nanowires based on the N-terminal region of the yeast prion, Sup35. Nanogold was covalently linked to the engineered cysteine residues in the protein and conjugate colloidal gold and silver particles were associated along the fibers to form wires,80 (B) assembly of diphenylalanine to form nanotubes that can be filled with silver to make nanowires.82
Conclusions
Amyloid fibrils are extremely strong, highly ordered and organized fibers that can be formed by a large number of proteins and peptides. Their stability and insolubility means that they are useful in a large number of naturally occurring forms as well as bionanotechnology. However, it also means that they are destructive and have the ability to accumulate in the tissues in disease. Although recent key advances have been made in understanding the structure of amyloid-like fibrils, better understanding of the mechanism of assembly is essential for combating amyloid diseases as well as exploiting the potential benefits of protein/peptide design.
Acknowledgements
R.N.R. is supported by a grant from the BBSRC, L.C.S. would like to acknowledge the support of the BBSRC, Alzheimer's research trust, Royal Society and Wellcome Trust. The authors would like to express their thanks to Dr. Vestergaard for kind contribution of Figure 3 and to Dr. Scheibel for Figure 4A and to Dr. Gazit for Figure 4B. They would also like to thank Dr. Curtis Rambaran for critical reading of the manuscript.
Note
This manuscript has been previously published: Rambaran RN, Serpell LC. Amyloid Fibrils: Abnormal Protein Assembly. In: Scheibel T, editor. Fibrous Proteins. Austin: Landes Bioscience; 2008. pp. 1–11.
Footnotes
Previously published online as a Prion E-publication: www.landesbioscience.com/journals/prion/article/7488
References
- 1.Pepys M. Pathogenesis, diagnosis and treatment of systemic amyloidosis. Phil Trans Soc Lond B. 2001;356:203–211. doi: 10.1098/rstb.2000.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fowler DM, Koulov AV, Alory-Jost C, et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedrich NaK A. Zur amyloidfrage. Arch Pathol Anat Physiol Klin Med. 1859;16:50–65. [Google Scholar]
- 4.Divry P, Florkin M. Sur les proprietes optiques de l'amyloid. Societe de Biologie. 1927;97:180–181. (Fre). [Google Scholar]
- 5.Cohen AS, Calkins E. Electron microscopic observation on a fibrous component in amyloid of diverse origins. Nature. 1959;183:1202–1203. doi: 10.1038/1831202a0. [DOI] [PubMed] [Google Scholar]
- 6.Cohen AS, Calkins E. The isolation of amyloid fibrils and a study of the effect of collagenase and hyaluronidase. J Cell Biol. 1964;21:481–486. doi: 10.1083/jcb.21.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 8.Glenner G, Ein D, Eanes E, et al. Creation of “Amyloid” Fibrils from Bence Jones Proteins. Science. 1971;174:712–714. doi: 10.1126/science.174.4010.712. [DOI] [PubMed] [Google Scholar]
- 9.Sunde M, Serpell L, Bartlam M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 10.Glenner GG, Terry W, Harada M, et al. Amyloid fibril proteins: proof of homology with immunoglobulin light chains by sequence analyses. Science. 1971;172:1150–1151. doi: 10.1126/science.172.3988.1150. [DOI] [PubMed] [Google Scholar]
- 11.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 12.Glenner GG, Wong CW. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 13.Botto M, Hawkins PN, Bickerstaff MC, et al. Amyloid deposition is delayed in mice with targeted deletion of the serum amyloid P component gene. Nat Med. 1997;3:855–859. doi: 10.1038/nm0897-855. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins PN, Myers MJ, Lavender JP, et al. Diagnostic radionuclide imaging of amyloid: biological targeting by circulating human serum amyloid P component. Lancet. 1988;1:1413–1418. doi: 10.1016/s0140-6736(88)92235-0. [DOI] [PubMed] [Google Scholar]
- 15.La Spada AR, Wilson EM, Lubahn DB, et al. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 16.Colon W, Kelly JW. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 17.Serpell L. Alzheimer's amyloid fibrils: structure and assembly. Biochim Biophys Acta. 2000;1502:16–30. doi: 10.1016/s0925-4439(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 18.McParland VJ, Kad NM, Kalverda AP, et al. Partially unfolded states of beta(2)-microglobulin and amyloid formation in vitro. Biochemistry. 2000;39:8735–8746. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 19.Jahn TR, Parker MJ, Homans SW, et al. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat Struct Mol Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 20.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nat Struct Mol Biol. 2006;13:202–208. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 21.Come J, Fraser P, Lansbury PJ. A Kinetic Model for Amyloid Formation In the Prion Diseases, Importance of Seeding. PNAS USA. 1993;90:5959–5963. doi: 10.1073/pnas.90.13.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomakin A, Chung DS, Benedek GB, et al. On the nucleation and growth of amyloid beta-protein fibrils: detection of nuclei and quantitation of rate constants. Proc Natl Acad Sci USA. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh DM, Hartley DM, Kusumoto Y, et al. Amyloid beta-protein fibrillogenesis. Structure and biological activity of protofibrillar intermediates. J Biol Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 24.Ding TT, Harper JD. Analysis of amyloid-beta assemblies using tapping mode atomic force microscopy under ambient conditions. In: Wetzel R, editor. Methods in Enzymology: Amyloid, prions and other protein aggregates. Vol 309. Academic press; 1999. pp. 510–525. [DOI] [PubMed] [Google Scholar]
- 25.Roher AE, Baudry J, Chaney MO, et al. Oligomerizaiton and fibril asssembly of the amyloid-beta protein. Biochim Biophys Acta. 2000;1502:31–43. doi: 10.1016/s0925-4439(00)00030-2. [DOI] [PubMed] [Google Scholar]
- 26.Harper JD, Wong SS, Lieber CM, et al. Assembly of A beta amyloid protofibrils: an in vitro model for a possible early event in Alzheimer‘s disease. Biochemistry. 1999;38:8972–8980. doi: 10.1021/bi9904149. [DOI] [PubMed] [Google Scholar]
- 27.Podlisny MB, Ostaszewski BL, Squazzo SL, et al. Aggregation of secreted amyloid beta-protein into sodium dodecyl sulfate-stable oligomers in cell culture. J Biol Chem. 1995;270:9564–9570. doi: 10.1074/jbc.270.16.9564. [DOI] [PubMed] [Google Scholar]
- 28.Lambert MP, Barlow AK, Chromy BA, et al. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harper JD, Wong SS, Lieber CM, et al. Observation of metastable Abeta amyloid protofibrils by atomic force microscopy. Chem Biol. 1997;4:119–125. doi: 10.1016/s1074-5521(97)90255-6. [DOI] [PubMed] [Google Scholar]
- 30.Perutz M, Johnson T, Suzuki M, et al. Glutamine Repeats as Polar Zippers: Their Possible Role in Inherited Neurodegenerative Diseases. PNAS USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlunegger MP, Bennett MJ, Eisenberg D. Oligomer formation by 3D domain swapping: a model for protein assembly and misassembly. Adv Protein Chem. 1997;50:61–122. doi: 10.1016/s0065-3233(08)60319-8. [DOI] [PubMed] [Google Scholar]
- 32.Staniforth RA, Giannini S, Higgins LD, et al. Three-dimensional domain swapping in the folded and molten-globule states of cystatins, an amyloid-forming structural superfamily. Embo J. 2001;20:4774–4781. doi: 10.1093/emboj/20.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 34.Malisauskas M, Zamotin V, Jass J, et al. Amyloid protofilaments from the calcium-binding protein equine lysozyme: formation of ring and linear structures depends on pH and metal ion concentration. J Mol Biol. 2003;330:879–890. doi: 10.1016/s0022-2836(03)00551-5. [DOI] [PubMed] [Google Scholar]
- 35.Bemporad F, Taddei N, Stefani M, et al. Assessing the role of aromatic residues in the amyloid aggregation of human muscle acyphophatase. Protein Science. 2006;15:862–870. doi: 10.1110/ps.051915806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De la Paz M, Serrano L. Sequence deteminants of amyloid fibril formation. Proc Natl Acad Sci USA. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DuBay KF, Pawar AP, Chiti F, et al. Prediction of the absolute aggregation rates of amyloidogenic polypeptide chains. J Mol Biol. 2004;341:1317–1326. doi: 10.1016/j.jmb.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 38.Pawar AP, Dubay KF, Zurdo J, et al. Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative disease. J Mol Biol. 2005;350:379–392. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Zibaee S, Makin OS, Goedert M, et al. A simple algorithm locates beta-strands in the amyloid fibril core of alpha-synuclein, Abeta and tau using the amino acid sequence alone. Protein Sci. 2007;16:906–918. doi: 10.1110/ps.062624507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, et al. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 41.Makin OS, Sikorski P, Serpell LC. Diffraction to study protein and peptide assemblies. Curr Opin Chem Biol. 2006;10:417–422. doi: 10.1016/j.cbpa.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Tycko R. Insights into the amyloid folding problem from solid-state NMR. Biochemistry. 2003;42:3151–3159. doi: 10.1021/bi027378p. [DOI] [PubMed] [Google Scholar]
- 43.Der-Sarkissian A, Jao CC, Chen J, et al. Structural organisation of alpha-synuclein studied by site-directed spin labelling. J Biol Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 44.Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem. 2004;279:48420–48425. doi: 10.1074/jbc.M406853200. [DOI] [PubMed] [Google Scholar]
- 45.Torok M, Milton S, Kayed R, et al. Structural and dynamic features of Alzheimer's Abeta peptide in amyloid fibrils studied by site-directed spin labelling. J Biol Chem. 2002;277:40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- 46.Goldsbury C, Kistler J, Aebi U, et al. Watching amyloid fibrils grow by time-lapse atomic force microscopy. J Mol Biol. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 47.Serpell L, Sunde M, Benson M, et al. The protofilament substructure of amyloid fibrils. J Mol Biol. 2000;300:1033–1039. doi: 10.1006/jmbi.2000.3908. [DOI] [PubMed] [Google Scholar]
- 48.Jimenez JL, Guijarro JI, Orlova E, et al. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jimenez JL, Tennent G, Pepys MB, et al. Structural diversity of ex vivo amyloid fibrils studied by cryo-electron microscopy. J Mol Biol. 2001;311:241–247. doi: 10.1006/jmbi.2001.4863. [DOI] [PubMed] [Google Scholar]
- 50.Jimenez JL, Nettleton EJ, Bouchard M, et al. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldsbury C, Kistler J, Aebi U, et al. Watching amyloid fibrils grow by time-lapse atomic force microscopy. J Mol Biol. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 52.Sachse C, Xu C, Wieligmann K, et al. Quaternary structure of a mature amyloid fibril from Alzheimer's Abeta(1–40) peptide. J Mol Biol. 2006;362:347–354. doi: 10.1016/j.jmb.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 53.Blake C, Serpell L. Synchrotron X-ray studies suggest that the core of the transthyretin amyloid fibril is a continuous b-helix. Structure. 1996;4:989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 54.Inouye H, Fraser P, Kirschner D. Structure of β-Crystallite Assemblies by Alzheimer Amyloid Protein Analogues: Analysis by X-Ray Diffraction. Biophys J. 1993;64:502–519. doi: 10.1016/S0006-3495(93)81393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geddes AJ, Parker KD, Atkins EDT, et al. “Cross β” Conformation in Protein. J Mol Biol. 1968;32:343–358. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- 56.Makin OS, Sikorski P, Serpell LC. Diffraction to study protein and peptide assemblies. Curr Opin Chem Biol. 2006;10:417–422. doi: 10.1016/j.cbpa.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 57.Makin OS, Atkins E, Sikorski P, et al. Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci USA. 2005;102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makin OS, Serpell LC. Structures for amyloid fibrils. FEBS J. 2005;272:5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- 59.Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 60.Sawaya MR, Sambashivan S, Nelson R, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 61.Tycko R. Progress towards a molecular-level structural understanding of amyloid fibrils. Curr Opin Struct Biol. 2004;14:96–103. doi: 10.1016/j.sbi.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Naito A, Kamihira M, Inoue R, et al. Structural diversity of amyloid fibril formed in human calcitonin as revealed by site-directed 13C solid-state NMR spectroscopy. Magn Reson Chem. 2004;42:247–257. doi: 10.1002/mrc.1323. [DOI] [PubMed] [Google Scholar]
- 63.Gordon DJ, Balbach JJ, Tycko R, et al. Increasing the amphiphilicity of an amyloidogenic peptide changes the beta-sheet structure in the fibrils from antiparallel to parallel. Biophys J. 2004;86:428–434. doi: 10.1016/S0006-3495(04)74119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balbach JJ, Petkova AT, Oyler NA, et al. Supramolecular structure in full-length Alzheimer's b-amyloid fibrils: evidence for parallel b-sheet organisation from solid state nuclear magnetic resonance. Biophys J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petkova AT, Buntkowsky G, Dyda F, et al. Solid state NMR reveals a pH-dependent anti-parallel beta-sheet registry in fibrils formed by a beta-amyloid peptide. J Mol Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 66.Petkova AT, Leapman RD, Guo Z, et al. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 67.Petkova AT, Ishii Y, Balbach JJ, et al. A structural model for Alzheimer's beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luhrs T, Ritter C, Adrian M, et al. 3D structure of Alzheimer's amyloid-beta(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vestergaard B, Groenning M, Roessle M, et al. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 2007;5:134. doi: 10.1371/journal.pbio.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer's disease. Trends Pharmacol Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 71.Lue LF, Kuo YM, Roher AE, et al. Soluble amyloid beta peptide concentration as a predictor of synaptic change in Alzheimer's disease. Am J Pathol. 1999;155:853–862. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naslund J, Haroutunian V, Mohs R, et al. Correlation between elevated levels of amyloid beta-peptide in the brain and cognitive decline. Jama. 2000;283:1571–1577. doi: 10.1001/jama.283.12.1571. [DOI] [PubMed] [Google Scholar]
- 73.Wang J, Dickson DW, Trojanowski JQ, et al. The levels of soluble versus insoluble brain Abeta distinguish Alzheimer's disease from normal and pathologic aging. Exp Neurol. 1999;158:328–337. doi: 10.1006/exnr.1999.7085. [DOI] [PubMed] [Google Scholar]
- 74.Hock C, Konietzko U, Papassotiropoulos A, et al. Generation of antibodies specific for beta-amyloid by vaccination of patients with Alzheimer's disease. Nature Medicine. 2002;8:0–5. doi: 10.1038/nm783. [DOI] [PubMed] [Google Scholar]
- 75.Smith JF, Knowles TP, Dobson CM, et al. Characterization of the nanoscale properties of individual amyloid fibrils. Proc Natl Acad Sci USA. 2006;103:15806–15811. doi: 10.1073/pnas.0604035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chapman MR, Robinson LS, Pinkner JS, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson LS, Ashman EM, Hultgren SJ, et al. Secretion of curli fiber subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sunde M, Kwan AH, Templeton MD, et al. Structural analysis of hydrophobins. Micron. 2007;39:773–784. doi: 10.1016/j.micron.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 79.Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol. 2002;56:703–741. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- 80.Scheibel T, Parthasarathy R, Sawicki G, et al. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc Natl Acad Sci USA. 2003;100:4527–4532. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baldwin AJ, Bader R, Christodoulou J, et al. Cytochrome display on amyloid fibrils. Journal of the American Chemical Society. 2006;128:2162–2163. doi: 10.1021/ja0565673. [DOI] [PubMed] [Google Scholar]
- 82.Reches M, Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 83.Serpell LC, Benson M, Liepnieks JJ, et al. Structural analyses of fibrinogen amyloid fibrils. Amyloid. 2007;14:199–203. doi: 10.1080/13506120701461111. [DOI] [PubMed] [Google Scholar]