Abstract
Exposure to high temperature or other stresses induces a synthesis of heat shock proteins. Many of these proteins are molecular chaperones and some of them help cells to cope with heat-induced denaturation and aggregation of other proteins. In the last decade, chaperones have received increased attention in connection with their role in maintenance and propagation of the Saccharomyces cerevisiae prions, infectious or heritable agents transmitted at the protein level. Recent data suggest that functioning of the chaperones in reactivation of heat-damaged proteins and in propagation of prions is based on the same molecular mechanisms but may lead to different consequences depending on the type of aggregate. In both cases the concerted and balanced action of “chaperones' team,” including Hsp104, Hsp70, Hsp40 and possibly other proteins, determines whether a misfolded protein is to be incorporated into an aggregate, rescued to the native state or targeted for degradation.
Key Words: Amyloid, Hsp40, Hsp70, Hsp104, stress response, yeast
Chaperones and Thermotolerance
The role of Hsps in development of induced thermotolerance.
A mild heat shock, which is nonlethal by itself, induces the synthesis of heat shock proteins (Hsps) and enhances the cell capacity to survive the subsequent severe heat shock exposure. This phenomenon is known as induced thermotolerance. It has been suggested that Hsp induction is essential for survival at elevated temperatures.1 A correlation has been found between the dynamics of development of induced thermotolerance in S. cerevisiae and synthesis of the heat shock protein with the molecular mass of about 100 kDa, later designated as Hsp104.2 Experiments with the deletion mutants clarified the role of Hsps in induced thermotolerance. Hsp104 is shown to play a crucial role in the induced thermotolerance in yeast,1,3,4 while Hsps of the Hsp70,4,5 Hsp40 and small Hsp6,7 families perform auxiliary functions in this process. Our focus on the Hsp104, 70 and 40, which are shown to cooperate with each other in disaggregation and refolding of the stress-damaged proteins.
General characteristics of molecular chaperones: “holdases,” “foldases” and “disagregases.”
Most Hsps are molecular chaperones facilitating protein folding, assembly and translocation across intracellular membranes. Roughly chaperones may be classified based on the mode of their interaction with substrate proteins. The ATP independent molecular chaperones, such as small heat shock proteins (sHsps) and proteins of Hsp40 family that can stabilize unfolded polypeptides but could not reactivate them are referred to as “holdases.”6,9 Some chaperones having ATP-binding domains that assist in folding of nonnative proteins via ATP-dependent binding and release are known as “foldases.” Proteins of the Hsp70 family also assist in folding, so that Hsp70s also seem to possess “foldase” activity. However, there is an alternative view considering the Hsp70 chaperones as “unfoldases” that use free energy from ATP binding and/or hydrolysis to unfold or pull apart misfolded and aggregated proteins to yield productive folding intermediates.8
The Hsp100/ClpB proteins are proposed to use ATP hydrolysis to disentangle aggregated polypeptides and transfer partially folded species to the Hsp70-Hsp40 binary system for subsequent refolding.4,10 In this context, Hsp100s act as “disaggregases.”11 However, Escherichia coli homologs of Hsp70 (DnaK) and Hsp40 (DnaJ) are thought to bind first to aggregated proteins, potentially helping Hsp100/ClpB to extract polypeptides from aggregates.10 Such an interplay is also shown to occur between sHsps and Hsp100/ClpB in yeast and bacteria.6,7,10
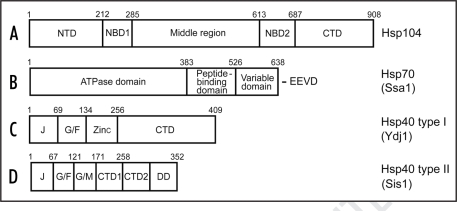
Yeast Hsp104. Hsp104 (Fig. 1A) is a yeast member of the Hsp100/ClpB family of AAA (ATPases associated with various cellular activities) superfamily of proteins, participating in a variety of cellular activities.10,11 In the presence of ATP, ADP or ATPγS, Hsp104 monomers are assembled into the hexamer complexes with an axial channel.12 Yeast Hsp104 has two nucleotide-binding domains, NBD1 and NBD2 with different catalytic properties.12,13 Mutations in NBD1 have little effect on hexamerization, while mutations in NBD2 severely impair hexamerization.14 There is an allosteric communication between NBD1 and NBD213 as well as communication between individual monomers in a hexamer, so that the ATP hydrolysis by Hsp104 is greatly influenced by hexamerization.15
Figure 1.
Structural organization of the yeast Hsp proteins involved in stress protection and prion propagation. (A) Hsp104; (B) Ssa1 as a representative of the Hsp70 family; (C) Hsp40 type I (Ydj1); (D) Hsp40 type II (Sis1). (The Hsp40 type III is not shown.) Designations: NBD, nucleotide-binding domain; NTD, N-terminal domain, middle region; CTD, C-terminal domain; J, J-domain; G/F, glycine and phenylalanine-rich region; Zinc, zinc-finger domain; G/M, glycine and methionine-rich region; DD, dimerization domain; E, glutamic acid; V, valine; D, aspartic acid. Numbers correspond to aa positions.
Hsp104 is induced by a mild heat shock treatment, and is crucial for induced thermotolerance in S. cerevisiae.1 It is also known to be induced in response to hydrogen peroxide,16 ethanol and sodium arsenite,1 and near-freezing cold shock.17 The ability of the hsp104Δ cells pretreated at 37°C to survive a lethal heat shock at 50°C is severely impaired but not completely abolished, indicating that other heat induced proteins also play role in this process.1 Specifically, Hsp70-Ssa and small heat shock protein Hsp26 become very important for thermotolerance in the S. cerevisiae cells lacking Hsp104.5,7 Aggregation of cellular proteins is a major consequence of severe stress, and Hsp104 is thought to act directly on protein aggregates, leading to their resolubilization.3 For efficient protein reactivation, Hsp104 requires the assistance of Hsp40 (Ydj1) and Hsp70 (Ssa).4
Abilities of Hsp104 to form a homohexamer and cooperatively bind and hydrolyze ATP are required for its functions in vivo.12,13 When ATP binding to NBD1 is impaired by a mutation, Hsp104 is unable to interact with substrates both in vitro and in vivo.18,19 Therefore, the ATP-bound state of NBD1 seems to be crucial for the chaperone-substrate interaction. Two models have been proposed for disaggregating action of Hsp104. In the first model, Hsp104 breaks up large aggregates into smaller ones in a crowbar-like activity. The second model suggests that a single polypeptide chain is extracted from an aggregate via translocation through the axial channel of Hsp104/ClpB hexamer, occurring by the unfolding/threading mechanism.10,11 The structure of the pore entrance of the Hsp104 oligomeric complex was shown to be crucial for Hsp104 function,20 supporting the latter mechanism.
Yeast Hsp70. All Hsp70 family proteins have three functionally separated domains: N-terminal 45 kDa ATP-binding domain, 15 kDa peptide-binding domain and C-terminal variable domain (Fig. 1B). Hsp70 transiently holds unfolded substrates in an intermediate state, preventing irreversible aggregation and catalyzing folding in the ATP dependent manner.21 However, the precise mechanism by which Hsp70 promotes folding is unclear thus far. There are at least two possible models of Hsp70's action, which are not mutually exclusive. Model 1 (‘kinetic partitioning’ model) suggests that Hsp70 plays a rather passive role. Via repetitive substrate binding and release cycles, it decreases the concentration of a free substrate. This prevents aggregation and allows more time for the substrate to fold into the native state. Model 2 (‘local unfolding’ model) proposes that Hsp70 induces local unfolding in the substrate, e.g., the untangling of a misfolded β-sheet, which helps to overcome kinetic barriers for folding to the native state. ATP energy may be needed either to induce such conformational changes or alternatively, to drive the ATPase cycle in the right direction.21 S. cerevisiae genome contains at least fourteen genes coding for the Hsp70 proteins. These proteins are localized in a variety of cellular compartments including the cytosol (subfamilies Ssa and Ssb), mitochondria (Ssc1 and Ssq1), endoplasmic reticulum (Kar2 and Lhs1), etc.22
Ssa subfamily is encoded by four genes: SSA1, 2, 3 and 4 (reviewed in ref. 22). SSA3 and SSA4 genes are expressed only at very low level in the exponentially growing cells, but are drastically induced after the temperature upshift, as well as by the stationary phase and other stresses. SSA2 is constitutively expressed, while SSA1 is normally expressed at moderate levels and induced by stresses. Deletion of any individual SSA gene does not affect induced thermotolerance. Double ssa1Δ ssa2Δ mutants grew slower than the parent at all temperatures and were unable to form colonies at 37°C, but their ability to induce tolerance to heat shock at 37°C was not changed. Moreover, double mutant displayed a higher level basal thermotolerance that is apparently due to upregulation of other Hsps.22,23 At least one of the Ssa proteins must be present to preserve the cell viability.22 Ssa proteins are implicated in protein translocation across intracellular membrane, prevention of aggregation of denatured proteins24 and cotranslational folding.25
Another yeast cytosolic Hsp70 family, Ssb, is not stress-inducible, and is encoded by two almost identical genes, SSB1 and SSB2. Strains with a single SSB gene disrupted exhibit no phenotypic change. However, a mutant with both genes disrupted grows slowly at all temperatures, and is cold sensitive.22 The major fraction of Ssb proteins has been found in association with the translating ribosomes, although some Ssb is distributed freely in the yeast cytosol. It is postulated that Ssb aids in folding of the emerging newly synthesized proteins.25 It is also possible that this protein is involved in protein turnover in the yeast by targeting misfolded proteins for degradation, as overexpression of Ssb1 suppressed the growth defects caused by some proteasome mutations.26
Yeast Hsp40 or J-Proteins. The Hsp40 family, subdivided into three subtypes, includes the structurally and functionally diverse proteins with one common feature, N-terminal J-domain (Fig. 1C and D).9 According to the current model,9,21 J-proteins first bind unfolded protein substrates in order to prevent their aggregation, and then transfer them to Hsp70, simultaneously stimulating the Hsp70 ATPase activity and thus stabilizing the Hsp70 interaction with the unfolded protein.
Type I Hsp40s, for instance yeast Ydj1, also contain the glycine and phenylalanine-rich (G/F) region, zinc finger-like domain, and conserved C-terminal domain (CTD). Type II Hsp40s, for example yeast Sis1, lack the zinc finger-like domain but contain extended glycine-rich region. The first 55 amino acids (aa) of this region are also rich in phenylalanines (G/F); the last 49 aa are rich in methionine residues (G/M). The C-terminal 181 aa of Sis1 contain the proposed polypeptide binding site (CTD1), a domain of unknown function (CTD2), and a dimerization domain.9,27
Both Ydj1 (type I) and Sis1 (type II) interact with Ssa, but not with Ssb. Ssb has its own Hsp40 cochaperone, zuotin or Zuo1.9 Ydj1 is not essential, but ydj1Δ deletion causes severe growth defects. In cooperation with Ssa, Ydj1 promotes the protein translocation across the intracellular membranes, and participates in refolding of the heat-damaged proteins. C-terminal domain of Ydj1 has been implicated in binding unfolded polypeptides.23 Apparently, zinc finger-like domain is necessary for transferring the nonnative polypeptides from Ydj1 to Hsp70.28 Ydj1 is also required for ubiquitin-dependent degradation of certain abnormal proteins.23
The essential protein Sis1 is shown to be less effective than Ydj1 in helping Ssa to suppress aggregation of stress damaged proteins but is linked to other processes, for example initiation of protein synthesis.25 Functions of Ydj1 and Sis1 are overlapping but not identical. Although excess Sis1 complements the slow growth phenotype of ydj1Δ, Ydj1 cannot complement the lethal phenotype of sis1Δ.24
Chaperone Effects on Prion Propagation
The role of Hsp104 in propagation of the yeast prions.
Yeast prions [PSI+], [PIN+] and [URE3] are self-perpetuating amyloid-like polymers of the proteins Sup35, Rnq1 and Ure2, respectively reviewed in refs. 29–31. Role of chaperones in prion propagation was first demonstrated for [PSI+].32 Search for genes that antagonize [PSI+] when present in the increased number of copies produced HSP104.33 Further investigation revealed that both overproduction and inactivation of Hsp104 cause loss of [PSI+].32 Surprisingly, overproduction of Hsp104 does not prevent de novo [PSI+] appearance.34,35 Hsp104 is also required for propagation of [PIN+]36 and [URE3],37 although high levels of Hsp104 do not antagonize these prions.
Recent experimental evidence38–41 (reviewed in ref. 29) supports a model35 postulating that the major role of Hsp104 in prion propagation in vivo is to break prion amyloids into smaller “seeds,” initiating new rounds of prion production (Fig. 2). This mechanism essentially means that disaggregating activity of Hsp104 converts amyloids into self-perpetuating prions.30,31 Variants of [PSI+] producing large aggregates that are relatively insensitive to Hsp104 require excess of this protein for efficient propagation.35,39
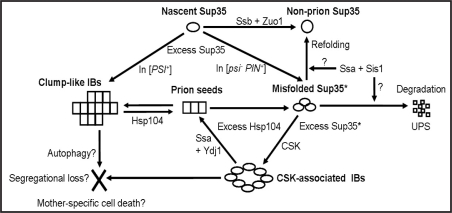
Figure 2.
Model for the role of molecular chaperones in formation and propagation of the [PSI+] prion. CSK, cytoskeleton; UPS, ubiquitin-proteasome system; IBs, inclusion bodies. See comments in text.
Dominant negative point mutations in either NBD (Fig. 1) disturb the Hsp104 ability to perform its role in both induced thermotolerance3,4,12,13 and prion propagation,13,32,38,42 indicating that both functions are associated with hexamerization and ATP hydrolysis. However, some mutant derivatives of Hsp104 that function efficiently in prion propagation can not protect yeast from extreme thermal stress.19 On the other hand, the dominant mutant derivative of Hsp104, bearing the A503V substitution in the middle region, increases size of the Sup35 aggregates in [PSI+] cells, leading to accumulation of cytologically detectable clumps and cytotoxicity, but decreases aggregate size and cytotoxicity of the polyglutamine fragment of human huntingtin expressed in yeast, and does not affect thermotolerance.43 A503V substitution impairs coordinated regulation of the NBD action without completely eliminating ATPase activity,44 that possibly leads to different consequences depending on with which type of aggregate that Hsp104 is interacting.
It remains unknown why does excess Hsp104 cause loss of [PSI+] but not of the other yeast prions. [PSI+] loss in the presence of excess Hsp104 probably requires other Hsp104 activities in addition to (or instead of) those involved in prion propagation, as N-terminal region of Hsp104 is required for [PSI+] curing by excess Hsp104 but not for [PSI+] maintenance.45 It is possible that excess Hsp104 solubilizes Sup35 prion polymers into monomers, that may require a mode of action distinct from one involved in oligomeric “seed” production. Alternatively, it was proposed that excess Hsp104 may impair prion segregation in cell divisions.46 Indeed, average size of remaining Sup35 polymers is increased in the presence of excess Hsp104,41 although this could be due to the fact that larger polymers are less sensitive to the Hsp104 disaggregating effect. The [PSI+] cells overexpressing Hsp104 accumulate cytologically detectable ring-like Sup35 structures that are not usually found in the [PSI+] cells with normal levels of Hsp104.34 Rings are also observed in the cells undergoing de novo [PSI+] induction in the presence of excess Sup35,34 and are shown to be associated with some components of the cortical actin cytoskeleton (CSK) involved in endocytosis, and/or with vacuolar membrane.47 It is possible that rings represent intermediates that arise in the process of either de novo prion formation or prion elimination, and are attempted by the cell to be targeted for elimination via autophagy and vacuolar proteolysis.
Effects of Other Chaperones on Prions and Polyglutamine Aggregates in Yeast
Ssa effects.
Increased levels of Ssa148 or any other member of Ssa subfamily49 enhanced phenotypic manifestation of [PSI+] and antagonized [PSI+] curing by excess Hsp104. Moreover, excess Ssa facilitated de novo [PSI+] induction in [psi−] cells by overproduced Sup35.49 Several semi-dominant mutations in the SSA1 gene have been obtained that decrease the mitotic stability of [PSI+].51,52 Strains with the mutant alleles of SSA1 were unable to propagate [PSI+] in the absence of the wild-type alleles of both SSA genes normally expressed in the absence of stress, SSA1 and SSA2.50 The second-site mutations in SSA1 restored normal prion propagation.51 Some (but not all) of the distantly related Hsp70 homologs from other organisms (plants and mammals) partly compensated the defect in [PSI+] maintenance observed in the presence of mutant Ssa.45 This shows that Ssa functions involved in [PSI+] maintenance are at least to a certain extent conserved in evolution.
Both calculations based on kinetics of [PSI+] curing,50 and biochemical assays such as size fractionation by chromatography and efficiency of fluorescence recovery after photobleaching indicated that size of the Sup35 aggregated structures is increased in the [PSI+] strains with mutant Ssa.53 It has been interpreted as an evidence of that Ssa helps to disassemble large aggregated structure into smaller polymers that become a target for the “shearing” action of Hsp104. On the other hand, Ssa overproduction slightly but reproducibly increased average size of prion polymers produced from large structures in the semi-denaturing conditions and visualized by semi-denaturing gel electrophoresis, that was also accompanied by an increase in the proportion of monomeric Sup35.49 As phenotypic manifestation of [PSI+] was enhanced rather than antagonized by excess Sup35, one could suggest that a significant fraction of the monomeric Sup35 generated in these cultures remained in the nonfunctional (possibly misfolded) state. Physical association between Ssa and Sup35 has been confirmed both in vivo and in vitro, suggesting that effects of Ssa on [PSI+] are due to its direct interaction with the prion-forming protein.49
While excess Ssa normally aids in propagation of the “conventional” variants of [PSI+], this effect could be reversed depending on the features of prion isolate and/or conditions affecting aggregate size and seed number. When Sup35 is overproduced in the [PSI+] strains, this leads to [PSI+] loss at low but detectable frequency, probably in result of accumulation of the large nontransmissible aggregates due to impairment of the balance between Sup35 and Hsp104.49 Notably, this effect is exacerbated in the presence of excess Ssa. Likewise, excess Ssa antagonizes propagation of the [PSI+] derivatives that are characterized by the abnormally large aggregate size and require increased levels of Hsp104 for their propagation.35,39
In contrast to its effect on conventional [PSI+], overproduction of Hsp70-Ssa1 impaired propagation of the yeast prion [URE3].54 Strangely enough, overproduction of the highly homologous Ssa2 protein did not show the same effect, and moreover, deletion of SSA2 impaired propagation of [URE3].55 Overproduction of some members of the Ssa subfamily counteracted poly-Q aggregation and/or toxicity in some yeast-based assays.43,56 However, at least in one genotypic background poly-Q aggregation was also decreased by double ssa1Δ ssa2Δ deletion.57
Ssb effects.
In a strong contrast to Ssa, Ssb proteins consistently act as [PSI+] antagonists. Excess Ssb increases [PSI+] curing by Hsp104 overproduction,58 inhibits [PSI+]-mediated suppression in certain [PSI+] isolates,58 and causes loss of [PSI+] upon prolonged incubation in certain genotypic backgrounds.59,60 Simultaneous deletion of both SSB1 and SSB2 genes decreases efficiency of [PSI+] curing by excess Hsp104 and increases the frequency of the spontaneous [PSI+] formation in [psi−] cells even in the absence of Sup35 overproduction.58 Ssb, like Ssa, can directly interact with Sup35.49 No effect of overproduced Ssb on poly-Q toxicity was detected.43,56
Difference between the Ssb and Ssa proteins in respect to [PSI+] curing is in significant part determined by their peptide-binding domains. The presence of Ssb peptide-binding domain is sufficient for an antiprion effect even when it is combined with the ATPase and variable domains of Ssa origin.49
Hsp40 effects.
Little is known about the effects of Hsp40 chaperones on yeast prions. Excess Ydj1 promoted loss of [URE3]37 and some variants of [PIN+],61 and somewhat antagonized the chimeric prion [PSI+]PS generated by the Sup35 protein with a prion domain from the distantly related yeast Pichia methanolica.62,63 The simultaneous overproduction of Ydj1 and Ssa1 cured some weak [PSI+] variants, but propagation of the strong variants remained unaffected.62 On the other hand, Ydj1 deficiency did not affect maintenance of [PIN+]64 or [PSI+]51 in the absence of other chaperone mutations. In the cells carrying the semi-dominant mutation SSA1-21, lack of Ydj1 further impaired mitotic stability of [PSI+].51
Another yeast cochaperone of the Hsp40 family, Sis1, appears to be required for propagation of the [PIN+] prion.64 Although Sis1 is an essential protein and therefore viable yeast cells lacking Sis1 cannot be constructed, in frame deletion within the SIS1 gene eliminated [PIN+], although it did not affect [PSI+]. In contrast, overproduction of Sis1 had no detectable effect on [PIN+] propagation.65 Sis1 is coprecipitated with the aggregated but not with the soluble form of Rnq1.64 An extended glycine-rich region of Sis1, including a region rich in phenylalanine residues (G/F) is critical for prion maintenance.64–66
Data on the effects of Hsp40 proteins on poly-Q aggregation in yeast are somewhat contradictory. In some assays, overproduction of Ydj1 counteracted aggregation of some poly-Q constructs in yeast.56 In the assay using prion-dependent poly-Q aggregation,57 Ydj1 and Sis1 exhibited opposite effects: excess Ydj1 increased the size and toxicity of poly-Q aggregates generated in the [PIN+] strain, while excess Sis1 decreased them.42 Likewise, mutation in the YDJ1 gene decreased poly-Q aggregation.57 Interestingly, these effects of Ydj1 and Sis1 somewhat parallel at least some observations made with their human homologs (Hdj2 and Hdj1, respectively) in the mammalian cells.67 Our preliminary data also indicate that overproduced Ydj1 and Sis1 proteins influence [PSI+] curing by excess Hsp104 in the opposite ways (S. Müller, J. Patterson and Y. Chernoff, unpublished).
Model for the chaperone effects.
Data reviewed above show that one and the same group of yeast chaperones is involved in protection against misfolded proteins and in prion propagation. We propose that effects of these chaperones on prion and nonprion aggregates are determined by one and the same molecular mechanism, while differences in effects are due to different parameters of aggregates. The model summarizing our current view is shown on Figure 2.
We propose that at normal levels, Hsp104 is responsible for propagation of the prion polymers via the subsequent cycles of breakage and growth. When the Hsp104/Sup35 ratio is shifted towards Sup35 in the [PSI+]-containing cells, polymer size is increased leading to the accumulation of large clump-shaped inclusion bodies (IBs) that are eliminated from the population either due to a segregation defect, followed by the death of IB-accumulating cells, or via autophagy. Abundance of such IBs is increased in the [PSI+] variants with the decreased sensitivity to Hsp104 (such as [PSI+] ≠ 104d)35 resulting in frequent loss of these variants in the normal conditions and their rescue at high levels of Hsp104.
Increase in the Hsp104 levels results in disruption of the ordered structure of prion aggregates. However, if overexpression of Hsp104 is not accompanied by the overexpression of the Hsp70 and Hsp40 chaperones, Hsp104 is not capable of solubilizing aggregates into the properly refolded monomers on its own. Therefore, disruption of the aggregate structure and increased hydrophobic exposure, induced by Hsp104, are followed by amorphous agglomeration of the misfolded prion protein molecules, rather than by solubilization. We propose that these misfolded agglomerated proteins can be recognized by Sis1, which in combination with Ssa, targets them for either refolding or degradation via the proteasomal pathway.
Alternatively, agglomerates are recognized by the CSK networks involved in endocytosis, including such proteins as Sla1, Sla2 and End3.47 This results in formation of the different kind of IBs, frequently having the ring-shaped morphology and associated with CSK. These IBs are intended to be targeted for degradation via autophagy and vacuolar pathway, but they can also be recognized by Ydj1, which in combination with Ssa stabilizes proteins in the misfolded state and promotes disassembly of IBs to oligomers, capable of reentering the prion propagation cycle. Therefore, Ssa effects depend on the participating cochaperones.
The Ssb protein is acting on the nascent Sup35 polypeptide, and its major function is to promote folding of the newly synthesized Sup35 into a nonprion form. In this way, Ssb is antagonizing prion formation and propagation, and its effects are additive to all other anti-prion factors as it is working in a separate pathway.
The same model also explains chaperone effects on de novo [PSI+] formation, as shown on Figure 2. Some parts of this model should certainly apply to the other prions as well, although other steps could turn out to be protein specific. E.g., effects of specific Hsps may vary depending on the prion, for example due to different affinities of the Hsp40s to different prion proteins. In case of polyglutamine aggregates lacking the prion propagating activity on their own, prevention of the aggregate degradation in the presence of excess Ydj1 leads to an increase in their size and toxicity.
Acknowledgements
This work was supported by grants R01GM58763 from NIH to Yury O. Chernoff and 04-04-48275a from Russian Foundation of Basic Research to Eugene G. Rikhvanov.
Note
This manuscript has been previously published: Rikhvanov EG, Romanova NV, Chernoff YO. Chaperone Effects on Prion and Nonprion Aggregates. In: Chernoff Y, editor. Protein-Based Inheritence. Austin and New York: Landes Bioscience and Kluwer Academic Press; 2007. pp. 83–92.
Footnotes
Previously published online as a Prion E-publication http://www.landesbioscience.com/journals/prion/article/5058
References
- 1.Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 2.McAlister L, Finkelstein DB. Heat shock proteins and thermal resistance in yeast. Biochem Biophys Res Commun. 1980;93:819–824. doi: 10.1016/0006-291x(80)91150-x. [DOI] [PubMed] [Google Scholar]
- 3.Parsell DA, Kowal AS, Singer M, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 4.Glover JR, Lindquist S. Hsp104, Hsp70 and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez Y, Parsell DA, Taulien J, Vogel JL, Craig EA, Lindquist S. Genetic evidence for a functional relationship between Hsp104 and Hsp70. J Bacteriol. 1993;175:6484–6491. doi: 10.1128/jb.175.20.6484-6491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: The structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 7.Cashikar AG, Duennwald ML, Lindquist SL. A chaperone pathway in protein disaggregation: Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005;280:23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slepenkov SV, Witt SN. The unfolding story of the Escherichia coli Hsp70 DnaK: Is DnaK a holdase or an unfoldase? Mol Microbiol. 2002;45:1197–1206. doi: 10.1046/j.1365-2958.2002.03093.x. [DOI] [PubMed] [Google Scholar]
- 9.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weibezahn J, Schlieker C, Tessarz P, Mogk A, Bukau B. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol Chem. 2005;386:739–744. doi: 10.1515/BC.2005.086. [DOI] [PubMed] [Google Scholar]
- 11.Bosl B, Grimminger V, Walter S. The molecular chaperone Hsp104 - A molecular machine for protein disaggregation. J Struct Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Parsell DA, Kowal AS, Lindquist SL. Saccharomyces cerevisiae Hsp104 protein: Purification and characterization of ATP-induced structural changes. J Biol Chem. 1994;269:4480–4487. [PubMed] [Google Scholar]
- 13.Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 2002;21:12–21. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schirmer EC, Queitsch C, Kowal AS, Parsell DA, Lindquist S. The ATPase activity of Hsp104, effects of environmental conditions and mutations. J Biol Chem. 1998;273:15546–15552. doi: 10.1074/jbc.273.25.15546. [DOI] [PubMed] [Google Scholar]
- 15.Schirmer EC, Ware DM, Queitsch C, Kowal AS, Lindquist SL. Subunit interactions influence the biochemical and biological properties of Hsp104. Proc Natl Acad Sci USA. 2001;98:914–919. doi: 10.1073/pnas.031568098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godon C, Lagniel G, Lee J, Buhler JM, Kieffer S, Perrot M, Boucherie H, Toledano MB, Labarre J. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 17.Kandror O, Bretschneider N, Kreydin E, Cavalieri D, Goldberg AL. Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol Cell. 2004;13:771–781. doi: 10.1016/s1097-2765(04)00148-0. [DOI] [PubMed] [Google Scholar]
- 18.Bosl B, Grimminger V, Walter S. Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J Biol Chem. 2005;280:38170–38176. doi: 10.1074/jbc.M506149200. [DOI] [PubMed] [Google Scholar]
- 19.Tkach JM, Glover JR. Amino acid substitutions in the C-terminal AAA+ module of Hsp104 prevent substrate recognition by disrupting oligomerization and cause high temperature inactivation. J Biol Chem. 2004;279:35692–35701. doi: 10.1074/jbc.M400782200. [DOI] [PubMed] [Google Scholar]
- 20.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 21.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craig EA, Gambill BD, Nelson RJ. Heat shock proteins: Molecular chaperones of protein biogenesis. Microbiol Rev. 1993;57:402–414. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson RJ, Heschl MF, Craig EA. Isolation and characterization of extragenic suppressors of mutations in the SSA hsp70 genes of Saccharomyces cerevisiae. Genetics. 1992;131:277–285. doi: 10.1093/genetics/131.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig EA, Eisenman HC, Hundley HA. Ribosome-tethered molecular chaperones: The first line of defense against protein misfolding? Curr Opin Microbiol. 2003;6:157–162. doi: 10.1016/s1369-5274(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 26.Ohba M. Modulation of intracellular protein degradation by SSB1-SIS1 chaperon system in yeast S. cerevisiae. FEBS Lett. 1997;409:307–311. doi: 10.1016/s0014-5793(97)00535-8. [DOI] [PubMed] [Google Scholar]
- 27.Sha B, Lee S, Cyr DM. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Sructure. 2000;8:799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 28.Fan CY, Ren HY, Lee P, Caplan AJ, Cyr DM. The type I Hsp40 zinc finger-like region is required for Hsp70 to capture nonnative polypeptides from Ydj1. J Biol Chem. 2005;280:695–702. doi: 10.1074/jbc.M410645200. [DOI] [PubMed] [Google Scholar]
- 29.Chernoff YO. Mutation processes at the protein level: Is Lamarck back? Mutat Res. 2001;488:39–64. doi: 10.1016/s1383-5742(00)00060-0. [DOI] [PubMed] [Google Scholar]
- 30.Chernoff YO. Replication vehicles of protein-based inheritance. Trends Biotechnol. 2004;22:549–552. doi: 10.1016/j.tibtech.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Chernoff YO. Amyloidogenic domains, prions and structural inheritance: Rudiments of early life or recent acquisition? Curr Opin Chem Biol. 2004;8:665–671. doi: 10.1016/j.cbpa.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 33.Chernoff YO, Ono B. Brown AJP, editor. Dosage-dependent modifiers of psi-dependent omnipotent suppression in yeast. (Protein Synthesis and Targeting in Yeast).NATO ASI. 1993;H71:101–110. [Google Scholar]
- 34.Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+] Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
- 35.Borchsenius AS, Müller S, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet. 2006;49:21–29. doi: 10.1007/s00294-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 36.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J. 2001;20:6683–6691. doi: 10.1093/emboj/20.23.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 42.Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 43.Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem. 2005;280:22809–22818. doi: 10.1074/jbc.M500390200. [DOI] [PubMed] [Google Scholar]
- 44.Schirmer EC, Homann OR, Kowal AS, Lindquist S. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol Biol Cell. 2004;15:2061–2072. doi: 10.1091/mbc.E02-08-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, Chernoff YO. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen KD, Wegrzyn RD, Chernova TA, Müller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: Novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones GW, Masison DC. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+] Genetics. 2003;163:495–506. doi: 10.1093/genetics/163.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones G, Song Y, Chung S, Masison DC. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones G, Song Y, Chung S, Masison DC. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot Cell. 2005;4:289–297. doi: 10.1128/EC.4.2.289-297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts BT, Moriyama H, Wickner RB. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast. 2004;21:107–117. doi: 10.1002/yea.1062. [DOI] [PubMed] [Google Scholar]
- 56.Muchowski PJ, Schaffar G, Sittler A, Wanker EE. Hayer-Hartl MK, Hartl FU, Hsp70 and Hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 60.Chacinska A, Szczesniak B, Kochneva-Pervukhova NV, Kushnirov VV, Ter-Avanesyan MD, Boguta M. Ssb1 chaperone is a [PSI+] prion-curing factor. Curr Genet. 2001;39:62–67. doi: 10.1007/s002940000180. [DOI] [PubMed] [Google Scholar]
- 61.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kushnirov VV, Kochneva-Pervukhova NV, Chechenova MB, Frolova NS, Ter-Avanesyan MD. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kryndushkin DS, Smirnov VN, Ter-Avanesyan MD, Kushnirov VV. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem. 2002;277:23702–23708. doi: 10.1074/jbc.M111547200. [DOI] [PubMed] [Google Scholar]
- 64.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aron R, Lopez N, Walter W, Craig EA, Johnson J. In vivo bipartite interaction between the Hsp40 Sis1 and Hsp70 in Saccharomyces cerevisiae. Genetics. 2005;169:1873–1882. doi: 10.1534/genetics.104.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyttenbach A, Carmichael J, Swartz J, Furlong RA, Narain Y, Rankin J, Rubinsztein DC. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington's disease. Proc Natl Acad Sci USA. 2000;97:2898–2903. doi: 10.1073/pnas.97.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]