Abstract
RNA silencing is a manifestation of a ubiquitous phenomenon that acts, at least in plants and some insects, as a natural defense mechanism against viruses. As a counter-strategy, viruses have evolved to encode silencing suppressor proteins (SSPs) that can block the defense response and evade the host immunity. Although numerous SSP have been identified, little information is available on the molecular basis of their mode of action. Among SSPs, the polerovirus protein P0 functions as an F-box protein that targets an essential actor of the silencing pathway. Our work demonstrates that one of the main targets is ARGONAUTE 1 (AGO1), a key component of the RISC effector complex. By a physical interaction with AGO1, P0 mediates AGO1 protein degradation in planta. This is the first report of a plant virus that exploits components of the host ubiquitination machinery to overcome RNA silencing. This unusual mode of action may provide some clues concerning the mechanism governing phloem tropism of poleroviruses.
Key words: PTGS, viral silencing suppressor, ARGONAUTE 1, F-box protein, proteolysis, arabidopsis, ubiquitination, viral phloem restriction
RNA silencing also known as RNA interference (RNAi) in animals, is a universal system in multicellular organisms in which RNA expression is specifically downregulated at the post-transcriptional level by a complex process involving small RNAs.1 Among these, the so-called small interfering RNAs (siRNAs) are 5′-phosphorylated molecules of 21–24 nucleotides (nt) generated by an RNase III-type Dicer enzyme from a double-stranded (ds) RNA. One strand of the siRNA can be incorporated into an RNA-induced silencing complex (RISC) to serve as a guide RNA to direct degradation of RNA species containing a complementary sequence. The core component of the RISC complex is an ARGONAUTE (AGO) protein, which contains the endonucleolytic cleavage activity. An important second class of small RNAs are the microRNAs (miRNAs) produced by Dicer from imperfect hairpin structures in noncoding transcripts of cellular origin. When incorporated into RISC they either direct cleavage (most commonly in plants) of transcripts bearing sequence homology or interfere with their translation.
In plants and invertebrates, virus infection is typically accompanied by the appearance of double-stranded RNAs and this can trigger a potent RNA silencing response aiming at degrading the foreign RNA. As a counter-strategy viruses encode silencing suppressor proteins (SSPs) that can block this host defense response.2 The molecular basis for suppressor activity has only been elucidated for a few of these proteins. For instance, the P19 protein encoded by Tomato Bushy Stunt Virus binds directly to siRNAs3,4 so as to impede their loading on RISC, their spread or their amplification by cellular RNA dependant RNA polymerases.5 Other SSPs, including the Closterovirus P21 and HC-Pro of Potyviruses can also bind siRNAs. This has led to the hypothesis that binding of ds siRNA may represent a widespread counter-strategy.6 Until recently, the only SSP that was known to function by interacting with a protein component of the silencing pathway was the 2b protein of Cucumber Mosaic Virus. By physically binding to AGO1, 2b provokes inhibition of the cleavage activity of the RISC machinery.7 More recent evidence now reveals that another SSP, the polerovirus P0 protein, has an even more dramatic impact on the fate of AGO1, possibly by hijacking components of the cellular ubiquitination pathway to promote degradation of AGO1.8,9
Poleroviruses are a group of plant viruses (Luteoviridae family) with a small plus sense-RNA genome. They encode a ∼29 kDa protein P0 which is required for strong viral pathogenesis.10 Pfeffer et al.11 demonstrated its silencing suppressor activity in a transient expression agroinfiltration assay using the GFP reporter gene. To investigate the mechanism of action of P0 a yeast two-hybrid screen of an Arabidopsis thaliana cDNA library was undertaken. Two cellular partners, SKP1 and SKP2, were shown to interact with P0 of two poleroviruses, Beet Western Yellows Virus (BWYV) and Cucurbit Aphid Borne Yellows Virus (CABYV).12 SKP proteins (S-phase Kinase-related protein) are subunits of the SCF (SKP-Cullin-F box) complex in the ubiquitin-dependent protein degradation pathway. The P0-SKP interaction was confirmed in vitro by pull-down assay and in planta by Bimolecular Fluorescence Complementation (BiFC) experiments. Moreover, a yeast bridging assay showed that CUL1 is part of a larger complex containing P0 and SKP, corroborating the existence of a novel SCFP0 complex. Recognition between P0 and SKP involves a domain present in different polerovirus P0's (which are otherwise rather dissimilar in sequence) that shows similarities with the consensus motif of F-box proteins. F-box proteins are responsible for substrate specificity of SCF complexes. Point mutation in the F-box motif of P0 abolished interaction with SKP, both in yeast and in planta, and concurrently diminished virus pathogenicity and the ability of P0 to suppress PTGS. Furthermore, plants in which SKP-homolog expression had been reduced by virus-induced gene silencing (VIGS) were resistant to polerovirus infection.12 These findings supported a model in which P0 functions as an F-box protein that directs the host SCF E3 ligase machinery to target a component of the PTGS pathway. This model thus established a link between the ubiquitination machinery and inhibition of the silencing pathway.
The second step in unraveling the mechanism of action of P0 was to identify its target. By using an inverted-repeat-PTGS agroinfiltration assay, we found that P0 had no effect on the biogenesis of primary siRNA but interfered with a step downstream of the action of DCL.9 Examination of transgenic plants expressing P0 revealed strong developmental abnormalities as well as enhanced accumulation of several miRNA-target transcripts, mimicking the effects observed in a hypomorphic ago1 mutant. These results were similar to those observed with the CMV 2b protein,7 suggesting that P0 may interfere with the RISC effector complex and target AGO1. Strikingly, plants expressing P0 showed a strong reduction of AGO1 protein level which was not correlated to a decrease of AGO1 transcript accumulation (in fact the levels increased, as expected since AGO1 transcript accumulation is under feedback control by a miRNA13). These findings provided strong circumstantial support for the hypothesis that P0 acts as an F-box protein to recognize AGO1 and promote its degradation (Fig. 1). Finally co-immunoprecipitation and BiFC experiments provided direct evidence for a physical interaction between P0 and AGO1 in vitro and in vivo.9
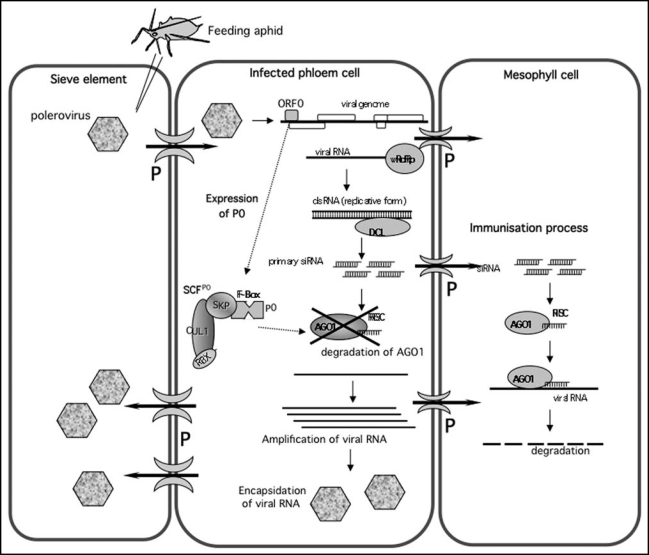
Figure 1.
Model of the process of infection by Poleroviruses. Poleroviruses are introduced into the phloem by feeding aphids having acquired virus on infected cells. The virus multiplies in companion and phloem parenchyma cells and circulates within the sieve tubes. In these cell types P0 inhibits RNA silencing by degrading AGO1. As siRNAs are not targeted by the virus, they may spread ahead of the infection front to the neighbouring cells (e.g., mesophyll cells) through the connecting plasmodesmata (P) and thereby immunize them against subsequent invasion by the virus. This immunisation process could be implicated in phloem restriction of poleroviruses. Encapsidation into virions would provide protection to the virus entering new phloem cells via the sieve tubes. vRdRp, virus-encoded RNA dependant polymerase; DCL, dicer-like protein; RISC, RNA Induced Silencing Complex; the SCFP0 complex contains an F-box protein (here P0), a SKP protein, a Cullin (CUL1) protein and a ring finger protein (RBX).
In a parallel study, Baumberger et al.,8 demonstrated AGO1 degradation by P0 in a transient expression assay. They localised the minimal sequence required for P0-mediated destabilisation of AGO1 to the PAZ domain (RNA binding domain) and the adjacent N-terminal domain. Tests performed to investigate the effect of proteasome inhibitors on AGO1 destabilisation were negative, which led Baumberger et al.,8 to suggest that the proteasome was not directly involved. A proposed alternative hypothesis could be that P0 behaves like a dominant negative inhibitor of a host F-box protein which may promote a specific pattern of ubiquitination of AGO1 required to fulfil its normal function. Thus, many questions remain to be answered concerning the role, if any, of ubiquitination and the proteasome in P0-induced AGO1 degradation.
In plants, RNA silencing is a non-cell-autonomous process.14 The silencing signal can spread to neighboring cells through plasmodesmata and for long distances via the vascular system. This obviously has implications for the plant's defense against virus infection in that it allows cells surrounding an infection site and in distant parts of the plant to be “primed” to resist infection before the virus arrives. The main silencing signal for localized movement has been shown to be the 21 nt siRNA produced by DCL4.15,16 The signal molecule(s) governing long-distance silencing remain to be identified, although, given the sequence-specific nature of the defense response, it will almost certainly prove to be a small RNA(s). Evidently, viruses that encode SSPs that interfere with siRNA production and/or sequester siRNAs should also interfere with movement of these silencing signals.2
Poleroviruses provide a particularly interesting model for the study of non-cell autonomous silencing signals because they are limited to the vascular tissues: the viruses multiply in companion cells and phloem parenchyma cells and circulate within the sieve elements to infect phloem cells in other leaves but they cannot invade mesophyll cells.17 Interestingly, using an agroinfiltration assay, Baumberger et al.,8 observed that P0 does not prevent the spread of the silencing signal to neighbouring cells. This is consistent with the finding that P0 does not interfere with the accumulation of siRNAs species but instead indirectly blocks their action at a downstream step. One intriguing possibility is that the movement of siRNAs from phloem cells into the surrounding mesophyll (and the presumed inability of the P0 produced in the infected phloem cells to move into these cells) “immunizes” these cells against subsequent invasion by the virus and thus plays a role in restricting poleroviruses to the phloem compartment (Fig. 1). A related question is, assuming that one or more of the small RNAs produced in the infected phloem cells are also involved in long-distance silencing via the sieve elements, how does the virus succeed in invading phloem cells in other parts of the plant? Do companion cells and phloem parenchyma cells respond differently than do mesophyll cells to non-cell-autonomous silencing signals? Does the virus specifically block the long-distance silencing pathway by a P0-independent mechanism? Exploration of these and other possibilities should not only shed light on the infection cycle of these interesting viruses but could also provide new insights into some of the plant-specific aspects of RNA silencing.
Acknowledgements
We would like to thank Véronique Brault for the design of the aphid.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5758
References
- 1.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 2.Li F, Ding SW. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 4.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding SW, Voinnet O. Antiviral Immunity directed by Small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, Lopez Moya JJ, Burgyan J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol. 2007;17:1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 9.Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol. 2007;17:1615–1621. doi: 10.1016/j.cub.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 10.Ziegler Graff V, Brault V, Mutterer JD, Simonis MT, Herrbach E, Guilley H, Richards KE, Jonard G. The coat protein of beet western yellows luteovirus is essential for systemic infection but the viral gene products P29 and P19 are dispensable for systemic infection and aphid transmission. Mol Plant Microb Interact. 1996;9:501–510. [Google Scholar]
- 11.Pfeffer S, Dunoyer P, Heim F, Richards KE, Jonard G, Ziegler Graff V. P0 of beet Western yellows virus is a suppressor of posttranscriptional gene silencing. J Virol. 2002;76:6815–6824. doi: 10.1128/JVI.76.13.6815-6824.2002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, Ziegler Graff V. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci USA. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 15.Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 17.Mayo MA, Ziegler Graff V. Molecular biology of luteoviruses. Adv Virus Res. 1996;46:413–460. doi: 10.1016/s0065-3527(08)60077-9. [DOI] [PubMed] [Google Scholar]