Abstract
Regulation of mRNA decay rates appears to be an important control point in determining the abundance of gene transcripts. Rapid change in decay rates of mRNAs could provide prompt responses of the plants to environmental fluctuations. SOS1 is a plasma-membrane Na+/H+ antiporter crucial for salt tolerance in Arabidopsis. In our recent paper in The Plant Journal, we have shown that SOS1 mRNA is inherently instable at normal growth conditions, but its stability is substantially elevated by salt and oxidative stress treatments. Salt stress-induced SOS1 mRNA stability is mediated by reactive oxygen species (ROS) produced, at least in part, through NADPH oxidases. We proposed a hypothetical model for the signaling pathway controlling SOS1 mRNA stability. In this addendum, we discuss the possible involvement of other components in conferring inherent instability and stress-induced stability of SOS1 mRNA.
Key words: mRNA stability, salt stress, signaling transduction, SOS1
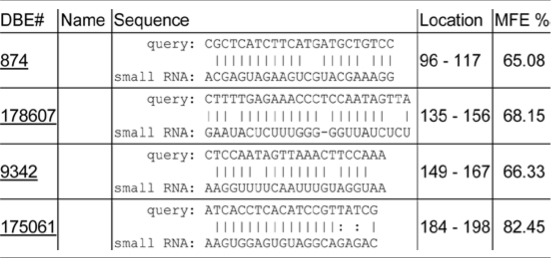
Genome-wide analysis of mRNA decay rates in Arabidopsis has indicated that mRNA half-lives vary from minutes to >24 hours.1 This research also concluded that transcripts that are miRNA targets generally have shorter half-lives.1 To determine if the inherent instability of SOS1 mRNA is attributed to mi- or si- RNA mediated mRNA degradation, we searched the Arabidopsis Small RNA Project database (http://asrp.cgrb.oregonstate.edu/db/)2 for possible small RNA target sequences in the 500 nt region of SOS1 mRNA shown essential for SOS1 mRNA instability. Although perfect complementary small RNA target sequences was not found in the 500 nt region, several hits with minimum free energy (MFE) over 60% were identified (Table 1). Mismatches between the small RNAs and the sequences in the 500 nt region raise the question as to whether these putative small RNAs could really direct degradation of SOS1 mRNA. Studies on recognition and regulation of target transcripts by miRNAs have suggested that ∼7 nt sites matching with the seed region at the 5′ end of the miRNA is important for target recognition and posttranscriptional gene repression.3–5 Mismatch studies revealed that efficiency of gene repression by miR-7 was strongly reduced by mismatches in positions 2 to 8. However, some of the mismatches between miR-278 and the target sequence in positions 2–7 still allowed repression of the reporter EGFP up to 50%. Even as few as four base pairs in positions 2–5 conferred efficient target regulation. Moreover, strong base pairing at 3′ end has compensatory role for insufficient 5′ end pairing for miRNA function.3 Thus, although mismatches appear between the target sequences and all four small RNAs, it is plausible that these four putative small RNAs could, at least at some extent, confer SOS1 mRNA instability.
Table 1.
Putative small RNA target sequences in the 500 nt region of SOS1 mRNA
 |
SOS1 mRNA stability was substantially increased upon salt treatment. Salt-induced SOS1 mRNA stability was impeded by the treatments using ROS scavengers and NADPH oxidase inhibitor. However, treatment using NADPH oxidase inhibitor DPI only partially inhibited salt-induced SOS1 mRNA accumulation, while treatments with ROS scavengers nearly abolished salt-induced SOS1 mRNA accumulation. These results suggested that the plasma membrane-bound NADPH oxidase might not be the sole source of ROS production mediating salt-induced SOS1 mRNA stability. Other possible sources for extracellular ROS production are pH-dependent cell wall peroxidases, germin-like oxalate oxidases and amine oxidases.6,7 These enzymes may also contribute to the stress induced apoplastic production of ROS that mediates salt-induced SOS1 mRNA stability. In addition to extracellular ROS, intracellular ROS produced in the mitochondria, chloroplast and peroxisome is also likely to play a role in salt-induced SOS1 mRNA stability. In fact, treatments with intracellular ROS-producing chemicals such as diuron, methyl viologen, menadione, or Rose Bengal induced SOS1 mRNA accumulation (data not shown). Diuron and methyl viologen are two chemicals that cause ROS production in chloroplast. Menadione causes ROS generation in mitochondria. Rose Bengal is a cell-permeable generator of singlet oxygen. Therefore, both extracelluar and intracellular ROS may play important role in mediating salt-induced SOS1 mRNA stability.
The intermediate signaling components transducing ROS signal to modulate SOS1 mRNA stability are remained to be identified. Since salt induction of SOS1 mRNA stability is a rapid process requiring only a few minutes, signal transduction events may solely occur in all existing molecules, including Ca2+ channels responsible for cytosolic Ca2+ elevation, membrane-bound NADPH oxidases, protein kinases and phosphotases and their substrate proteins, and the mRNAs to be stabilized. RNA binding proteins might be also involved. In yeast, RNA-binding protein Csx1 stabilizes the transcripts of the MAPK-regulated transcription factor Atf1 and mediates global control of gene expression in response to oxidative stress.8 In Arabidopsis, MAPK signaling cascade is modulated by ROS in response to various biotic and abiotic stresses.9 Several Csx1 homologs in Arabidopsis have been identified by sequence similarity search. The involvement of MAPKs and Csx1 homologous RNA-binding proteins in salt-induced SOS1 mRNA stability is currently under investigation.
Acknowledgements
This work was supported by the US Department of Agriculture National Research Initiative competitive grants 2004-35100-14863 and 2007-35100-18378 to H.Shi.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5821
References
- 1.Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD. ASRP: The Arabidopsis small RNA project database. Nucl Acid Res. 2005;33:637–640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Grimson A, Farh KK, Johnston WK, Garrett Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 7.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez Gabriel MA, Burns G, McDonald WH, Martín V, Yates JR, 3rd, Bähler J, Russell P. RNA-binding protein Csx1 mediates global control of gene expression in response to oxidative stress. EMBO J. 2003;22:6256–6266. doi: 10.1093/emboj/cdg597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitzschke A, Hirt H. Mitogen-activated protein kinases and reactive oxygen species signaling in plants. Plant Physiol. 2006;141:351–356. doi: 10.1104/pp.106.079160. [DOI] [PMC free article] [PubMed] [Google Scholar]


