Abstract
Resistance (R) gene-mediated immunity provides plants with rapid and strain-specific protection against pathogen infection. Our recent study using the genetically tractable Arabidopsis and turnip crinkle virus (TCV) pathosystem revealed a novel component, named CRT1 (compromised for recognition of the TCV CP), that is involved in general R gene-mediated signaling, including that mediated by HRT, an R gene against TCV. The Arabidopsis CRT1 gene family contains six additional members, of which two share high homology to CRT1 (75 and 81% a.a. identity); either CRT1 or its closest homolog restore the cell death phenotype suppressed by crt1. Analysis of single knock-out mutants for CRT1 and its closest homologs suggest that each may have unique and redundant functions. Here, we provide insight into the screening conditions that enabled identification of a mutant gene despite the presence of functionally redundant family members. We also discuss a potential mechanism that may regulate the interaction between CRT1 and R proteins.
Key words: resistance gene, ATPase, suppressor screening, Arabidopsis, turnip crinkle virus
Plant resistance (R) proteins activate defense signaling pathways following detection of a specific pathogen-encoded effector, or perception that a host factor has been altered by a pathogen effector. The vast majority of R proteins contain nucleotide binding site (NBS) and leucine-rich repeat (LRR) domains. These R proteins can be further divided into two subgroups, TIR-NBS-LRR and CC-NBS-LRR, depending on whether the N terminus consists of a Toll-interleukin 1 receptor (TIR) or a coiled-coiled (CC) domain, respectively.1 Subsequent to pathogen perception, the signal(s) generated by various R proteins likely converge into a limited set of pathways, with CC-NBS-LRR proteins usually utilizing NDR1 and TIR-NBS-LRR proteins generally requiring EDS1.2 However, the molecular mechanism(s) through which R proteins recognize a pathogen(s) and initiate a defense signal(s) remains unclear.
To gain insights into this elusive signaling process, several groups have performed genetic screens to isolate mutants whose R gene-mediated resistance responses are suppressed following either pathogen infection or expression of a transgene-encoded bacterial effector protein. Several proteins, including HSP90, SGT1 and RAR1, were shown to be required for resistance triggered by a variety of R proteins, suggesting their universal function in R protein-mediated resistance.3–7 However, while some R protein-mediated signaling pathways required both RAR1 and SGT1, others needed only one or neither protein. Thus, the requirement for RAR1 and SGT1 appears to be specific to each pathway.8 Further studies revealed that SGT1, RAR1 and HSP90 regulate the stability/accumulation of various R proteins,8–11 raising the possibility that they serve as (co)chaperones for assembling an active R protein complex.
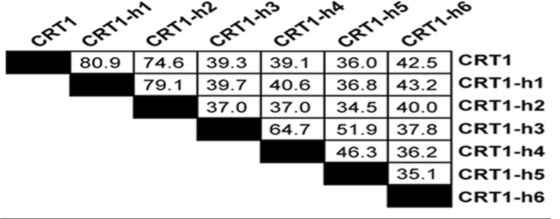
The Arabidopsis R protein HRT was previously shown to recognize the coat protein (CP) of turnip crinkle virus (TCV) and trigger necrotic lesion formation in the inoculated leaf, as well as local and systemic defense responses.12 To identify components of the HRT-mediated signaling pathway, a line containing HRT and an inducible CP transgene was constructed and screened for suppressors of CP-induced cell death.13 One mutant, named crt1 (compromised for recognition of the TCV CP), was identified; it contains a mutation in a GHKL (Gyrase, Hsp90, histidine kinase, MutL) ATPase.13 Interestingly, HSP90 also belongs to this recently recognized ATPase superfamily, although sequence homology between HSP90 and CRT1 is limited to the ATPase domain.14 Either wt CRT1 or its closest homolog, CRT1-h1 (81% a.a. identity to CRT1; Table 1), restores CP-induced cell death phenotype in the crt1 background,13 suggesting that CRT1 and CRT1-h1 are functionally redundant.
Table 1.
Amino-acid sequence identity between CRTI family members in Arabidopsis
 |
Given the presence of a functionally redundant homolog sharing 81% a.a. identity to CRT1, it is surprising that the crt1 mutant was identified. Because a previous study using the dexamethasone inducible system reported severe growth arrest and induction of defense-related genes when any transgene was highly expressed,15 we started with a transgenic line expressing CP at a level that was low (particularly in comparison to those attained during TCV infection), yet was sufficient to trigger cell death in non-mutant plants. The low level of CP expression in our transgenic line may have inadvertently provided screening conditions under which a rather modest compromise in R protein-mediated signaling could be detected, such as a mutation in a gene with functionally redundant family members. The crt1 and other crt mutants indeed showed cell death when CP was highly expressed via TCV infection. Thus, it is likely that crt1 would have escaped the suppressor screen if expression of the CP transgene had been higher. Another anti-viral R protein of Arabidopsis, RCY1, was utilized for a similar suppressor screen except that the effector protein was provided via viral infection.16 This screen identified mutations only in RCY1, consistent with our hypothesis that weak activation of the defense signaling pathway facilitated detection of a mutation in a gene that is part of a functionally redundant family.
Since HRT-mediated resistance to TCV was impaired in crt1 and was further compromised by silencing closely related CRT1 family members,13 the functional copy number of CRT1 family members appears to be important for resistance. This result, combined with the semi-dominant nature of the crt1 mutation led us to test whether the mutant phenotype is due to haploid insufficiency. Analysis of single T-DNA knockout mutants for CRT1 or CRT1-h1 revealed that resistance to Pseudomonas syringae was not compromised, although it was suppressed in a double knockout mutant (unpublished). These results suggest that loss of a single copy of CRT1 is not sufficient to compromise TCV resistance signaling, thereby arguing that the crt1 phenotype is due to a dosage effect of disabled CRT1 family members. An alternative, although mutually not exclusive, possibility is that crt1 suppresses TCV resistance via a negative gain of function. Ectopic expression of some truncated CRT1 variants suppressed cell death triggered by the constitutively activated R protein ssi4.13 Thus, crt1 might suppress resistance signaling by competing with wild type CRT1 for an interacting partner, likely an R protein. Such a scenario could explain why CRT1 dosage affects TCV resistance.
An intriguing possibility raised in a preview to our paper is that CRT1 may activate/prime a cytosolic R protein, which is then localized to the nucleus.17 Several lines of evidence suggest that nuclear localization of some R proteins is required for their function.18–20 Thus, CRT1 could be an important player that transits R proteins from one subcellular location to another, although it remains to be demonstrated whether HRT and the other R proteins shown to interact with CRT1 change subcellular location during resistance signaling. Another important question is what triggers CRT1 to activate/prime a client R protein. Western analysis has revealed that CRT1 is present as two distinct isoforms; the larger isoform presumably is created by an unknown post-translational modification.13 Interestingly, the larger CRT1 isoform interacts poorly with the NBS domain of HRT,13 suggesting that this putative modification is a crucial signal to release a client R protein. Thus, characterization of this post-translational modification may provide crucial insight into an R protein-mediate signaling pathway(s) that has been enigmatic for over a decade.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/5822
References
- 1.Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 2.Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tor M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Turk F, Can C, Dangl JL, Holub EB. Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell. 2002;14:993–1003. doi: 10.1105/tpc.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muskett PR, Kahn K, Austin MJ, Moisan LJ, Sadanandom A, Shirasu K, Jones JD, Parker JE. Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell. 2002;14:979–992. doi: 10.1105/tpc.001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tornero P, Dangl JL. A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 2001;28:475–481. doi: 10.1046/j.1365-313x.2001.01136.x. [DOI] [PubMed] [Google Scholar]
- 7.Tornero P, Merritt P, Sadanandom A, Shirasu K, Innes RW, Dangl JL. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell. 2002;14:1005–1015. doi: 10.1105/tpc.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt BF, 3rd, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 9.Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, Sadanandom A, Casais C, Parker J, Shirasu K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006;25:2007–2016. doi: 10.1038/sj.emboj.7601084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieri S, Mauch S, Shen QH, Peart J, Devoto A, Casais C, Ceron F, Schulze S, Steinbiss HH, Shirasu K, Schulze Lefert P. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16:3480–3495. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooley MB, Pathirana S, Wu HJ, Kachroo P, Klessig DF. Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell. 2000;12:663–676. doi: 10.1105/tpc.12.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang HG, Kuhl JC, Kachroo P, Klessig DF. CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to Turnip Crinkle Virus. Cell Host Microbe. 2008;3:48–57. doi: 10.1016/j.chom.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 15.Kang HG, Fang Y, Singh KB. A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J. 1999;20:127–133. doi: 10.1046/j.1365-313x.1999.00575.x. [DOI] [PubMed] [Google Scholar]
- 16.Sekine KT, Ishihara T, Hase S, Kusano T, Shah J, Takahashi H. Single amino acid alterations in Arabidopsis thaliana RCY1 compromise resistance to Cucumber mosaic virus, but differentially suppress hypersensitive response-like cell death. Plant Mol Biol. 2006;62:669–682. doi: 10.1007/s11103-006-9048-4. [DOI] [PubMed] [Google Scholar]
- 17.Monaghan J, Li X. R protein activation: another player revealed. Cell Host Microbe. 2008;3:9–10. doi: 10.1016/j.chom.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 19.Burch Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]


