Abstract
Lysine acetylation and its regulatory enzymes are known to have pivotal roles in mammalian cellular physiology. However, the extent and function of this modification in prokaryotic cells remain largely unexplored, thereby presenting a hurdle to further functional study of this modification in prokaryotic systems. Here we report the first global screening of lysine acetylation, identifying 138 modification sites in 91 proteins from Escherichia coli. None of the proteins has been previously associated with this modification. Among the identified proteins are transcriptional regulators, as well as others with diverse functions. Interestingly, more than 70% of the acetylated proteins are metabolic enzymes and translation regulators, suggesting an intimate link of this modification to energy metabolism. The new dataset suggests that lysine acetylation could be abundant in prokaryotic cells. In addition, these results also imply that functions of lysine acetylation beyond regulation of gene expression are evolutionarily conserved from bacteria to mammals. Furthermore, we demonstrate that bacterial lysine acetylation is regulated in response to stress stimuli.
Lysine acetylation is a dynamic, reversible, and regulatory post-translational modification in mammalian cells. Lysine acetylation status and its regulatory enzymes have been shown to influence several fundamental cellular pathways in mammalian cells, including cell survival and apoptosis, cellular differentiation, and metabolism. Dysregulation of the modification is associated with aging (1) and a few diseases, such as cancer, cardiovascular diseases, and neurodegenerative diseases (1–5).
Emerging evidence suggests diverse non-nuclear roles of lysine acetylation and its regulatory enzymes, in addition to its well recognized functions in DNA-templated processes. For example, Sirt1 modulates diverse cellular processes, such as fat metabolism, insulin production, glucose homeostasis, and cell survival, of which some are mediated through non-nuclear proteins (6–8). Three members of the sirtuin family (Sirt3, Sirt4, and Sirt5) are located in the mitochondrion (9). Activation of Sirt1, a mammalian ortholog of yeast Sir2, by resveratrol leads to diverse metabolic changes in animals (10). The roles of lysine acetylation in metabolism are further exemplified by the facts that the modification is present in more than 20% of mitochondrial proteins and is highly enriched among metabolic enzymes (11–14). Although the inventory and biological functions of lysine acetylation substrates in eukaryotic cells have begun to unfold, especially in histone proteins and transcription factors (15–19), the nature of lysine acetylation substrates in prokaryotic cells remains largely unknown.
The high abundance of lysine acetylation in mammalian mitochondrial proteins implies the possible widespread existence of the modification in prokaryotes, given the evolutionary lineage of eukaryotic mitochondria from bacteria (20). Acetyl-coenzyme A (CoA) synthetase, CheY, and Alba are the only proteins in prokaryotes known to be lysine-acetylated (4, 21–24). In Salmonella enterica, the lysine acetylation status of acetyl-CoA synthetase is regulated by CobB deacetylase, a Sir2 homolog in bacteria (21, 26), as well as Pat acetyltransferase (27). Acetyl-CoA synthetase activation by CobB sirtuin deacetylation is required for the bacteria to grow on short-chain fatty acids such as acetate and propionate. Interestingly, both human Sirt2 and yeast Sir2 proteins could restore growth of CobB sirtuin-deficient strains of S. enterica on short-chain fatty acids, suggesting that the sirtuins may have evolutionarily conserved roles in cellular metabolism (28). Despite evidence of the presence and roles of lysine acetylation in prokaryotes, the extent of lysine acetylation has not been carefully examined before.
Here we report the first global analysis of lysine acetylation in Escherichia coli. The proteomics study involves efficient affinity enrichment of lysine-acetylated, tryptic peptides with anti-acetyllysine antibodies and subsequent peptide identification for nano-HPLC/mass spectrometric analysis. The screening identified 138 lysine acetylation sites in 91 proteins in E. coli, of which 25% had mammalian orthologues. Our results suggest that diverse groups of bacterial proteins are the substrates of lysine acetylation, including metabolic enzymes, stress response proteins, and transcription and translation factors. The lysine acetylation substrates in E. coli are highly enriched in metabolic enzymes (∼53%) and proteins involved in translation (∼22%), two processes that are intimately linked to cellular energy status. These data therefore reveal previously unappreciated roles of lysine acetylation in the regulation of prokaryotic biochemical pathways and imply that DNA-independent functions of lysine acetylation are evolutionarily conserved from prokaryotic to eukaryotic cells.
EXPERIMENTAL PROCEDURES
Materials—
The reagents used in this work include Protein A-conjugated-agarose beads from Amersham Biosciences; Luria-Bertani (LB) medium from Invitrogen; iodoacetamide, C18 ziptips from Millipore Corp. (Bedford, MA); Luna C18 resin from Phenomenex (Torrance, CA). E. coli strains MG1655 and JW1106 were both acquired from the Coli Genetic Resource Center at Yale University. MG1655 is the wild type, and JW1106 is a CobB-deficient single-gene knockout of the Keio Collection (29).
Two anti-acetyllysine antibodies were used: an affinity-purified anti-acetyllysine polyclonal antibody from ImmuneChem Pharmaceuticals Inc. (Burnaby, British Columbia, Canada) and an anti-acetyllysine monoclonal antibody from Cell Signaling Technology (Boston, MA). Because the two antibodies were generated using different antigens (acetyllysine peptide library or acetyllysine residue, please see the vendor information for details) and purified differently (30, 31), they would likely have different binding specificities. Accordingly, a more diverse panel of acetyllyisne substrate peptides would be identified by using two antibodies for the described proteomics screening.
Preparation of Cell Lysate from E. coli—
E. coli DH5 was grown aerobically in LB medium at 37 °C. The cultured cells were harvested during the exponential growth phase by centrifugation at 4500 × g for 10 min and washed twice by resuspension of the pellet in ice-cold phosphate-buffered saline buffer (0.1 m Na2HPO4, 0.15 m NaCl, pH 7.2). The cells were resuspended in chilled lysis buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm dithiothreitol) and then sonicated with 12 short bursts of 10 s followed by intervals of 30 s for cooling. Unbroken cells and debris were removed by centrifugation at 4 °C for 30 min at 21,000 × g. The supernatant was divided into aliquots and stored at −80 °C until use.
In-solution Tryptic Digestion—
Five milligrams of proteins were precipitated with acetone, followed by centrifugation at 22,000 × g for 10 min. The resulting pellet was digested according to a previously described procedure (32). The protein pellet was rinsed twice with cold acetone to remove residual salts, resuspended in 50 mm NH4HCO3 (pH 8.5) (the protein pellet was not completely re-dissolved but rather was suspended as small particles), and digested with trypsin (Promega, Madison, WI) at an enzyme-to-substrate ratio of 1:50 for 16 h at 37 °C to enhance the solubility of the proteins prior to reduction and alkylation. The tryptic peptides were reduced with 5 mm dithiothreitol at 50 °C for 30 min and then alkylated using 15 mm iodoacetamide at ambient temperature for 30 min in darkness. The reaction was terminated with 15 mm cysteine at ambient temperature for 30 min. To ensure complete digestion, additional trypsin at an enzyme-to-substrate ratio of 1:100 was added to the peptide mixture, and the mixture was incubated for an additional 3 h.
Affinity Purification of Lysine-acetylated Peptides—
The anti-acetyllysine antibodies from ImmuneChem Pharmaceuticals Inc. and Cell Signaling Technology were mixed at a ratio of 1:1 and then immobilized on protein A-conjugated-agarose beads at 4–6 mg/ml by incubation at 4 °C for 4 h. The supernatant was removed, and the beads were washed three times with NETN buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40).
The tryptic peptides obtained from in-solution digestion were re-dissolved in NETN buffer. Insoluble particles were removed by centrifugation. Affinity purification was carried out by incubating the peptides with 20 μl of anti-acetyllysine antibody protein A-immobilized-agarose beads at 4 °C for 6 h with gentle shaking. The beads were washed three times with 1 ml of NETN buffer and twice with ETN (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA). The bound peptides were eluted from the beads by washing three times with 50 μl of 1% trifluoroacetic acid. The eluates were combined and dried in a SpeedVac. The resulting peptides were cleaned with C18 ZipTips (Millipore Corp.) according to the manufacturer's instructions, prior to nano-HPLC/mass spectrometric analysis.
HPLC/MS/MS Analysis—
HPLC/MS/MS analysis was performed in an integrated system that includes an Agilent 1100 series nanoflow liquid chromatography system (Agilent, Palo Alto, CA) and an LTQ two-dimensional trap mass spectrometer (Thermo Electron, Waltham, MA) equipped with a nanoelectrospray ionization source. One μl of tryptic peptides in buffer A (97.95% water/2% acetonitrile/0.05% acetic acid) was manually injected and separated in a capillary HPLC column (11 cm length × 75 μm inner diameter) packed in-house with Luna C18 resin (5 μm particle size, 100 Å pore diameter) (Phenomenex). Peptides were eluted from the column with a gradient of 6.0% to 90% buffer B (90% acetonitrile/9.95% water/0.05% acetic acid) in a 2 h LC/MS/MS analysis. The eluted peptides were electrosprayed directly into the LTQ ion trap mass spectrometer. Liquid chromatography tandem mass spectrometry was operated in a data-dependent mode such that the ten strongest ions in each MS scan were subjected to collisionally activated dissociation with a normalized collisionally activated dissociation energy of 35%.
Protein Sequence Database Search and Manual Verification—
Tandem mass spectra were used to search the E. coli entries (51,059 E. coli sequences) of the NCBI-nr database (updated July 31th, 2006 with a total of 3,841,279 sequences). Only the E. coli subset of the database was used for search because we are only interested in E. coli acetylation in the current study and all the cell lysate was from E. coli. The search engine MASCOT (version 2.1, Matrix Science, London, UK) was used for database search, and extract_msn.exe version 4.0 was used for peaklist generation. A low cutoff of peptide score 20 was selected to maximize the identification of lysine-acetylated peptides. Trypsin was specified as the proteolytic enzyme, and up to 6 missed cleavage sites per peptide were allowed. Carbamidomethylation of cysteine was set as a fixed modification and oxidation of methionine and acetylation of lysine as variable modifications. Charge states of +1, +2, or +3 were considered for parent ions. Mass tolerance was set to ±4.0 Da for parent ion masses and ± 0.6 Da for fragment ion masses. Acetylated lysine containing peptides identified with a MASCOT score of 25 were manually verified by the method described previously (33).
Western Blotting Analysis—
E. coli MG1655 and JW1106 were grown, harvested, and lysed as described above. Protein concentration was determined using the Bradford assay (Bio-Rad). Forty μg of proteins from E. coli cells was resolved by 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% milk at ambient temperature for 1 h. Then the membrane was incubated with anti-acetyllysine monoclonal antibody (0.8 μg/ml in TBST (25 mm Tris-HCl, pH 8.0 125 mm NaCl, 0.1% Tween 20) with 3% BSA1; from Cell Signaling Technology) overnight at 4 °C. After washing with TBST four times for 5 min each, the membrane was incubated with horseradish peroxidase-conjugated anti-mouse IgG (1 μg/ml in TBST with 3% BSA) at ambient temperature for 2 h. The ECL system (PerkinElmer Life Sciences) was used for signal detection. To carry out the competition experiment, the anti-acetylated lysine antibody was pre-incubated with 3% BSA (acetylated or non-acetylated) at ambient temperature for 2 h before it was incubated with the membrane.
Hypoxic Treatment of E. coli—
Hypoxic treatment of E. coli was carried out as described previously (34). E. coli MG1655 was grown in LB medium in 125-ml flasks. When its optical density (A600) at 600 nm reached 0.3, it was exposed to one of the following conditions: nitrogen only, 5% or 25% air. For these three experiments, nitrogen was bubbled through the cell suspension for 15 min, and the flasks were sealed tightly immediately. For the latter two experiments, 5% or 25% air was then introduced back to the flasks by a syringe needle, and further sealing was applied immediately. The cells were allowed to continue to grow until their A600 reached 0.6. The cells were harvested and prepared for Western blotting analysis as described above.
Starvation of E. coli—
Starvation of E. coli was carried out as described (35). E. coli MG1655 was grown in LB medium as described above. When its A600 reached 0.3, starvation was imposed by sudden depletion of all carbon sources as follows: cells were centrifuged at 4500 × g for 10 min at 4 °C and washed twice with two volumes of sterile ice-cold M9 minimal medium (no carbon source). The cells were resuspended in the same volume of M9 minimal medium and incubated overnight. The cells were harvested and prepared for Western blotting analysis as described above.
Structure Analysis of E. coli Lysine Acetylation Sites—
For each acetylated E. coli protein, BLAST was run against a database of domain sequences with known structures from the SCOP90 representative set of ASTRAL compendium (version 1.71) (36, 37). Database size of BLAST was set to the size of the protein nr database as of April 6th, 2007 (39,280,211,952 letters) to impose a stringent E-value cutoff. Hits with an E-value less than 0.001 were analyzed. We identified homologous structures for 69 of the 91 acetylated proteins (∼76%). For these proteins, we mapped the positions of acetylated lysines to the model structures using BLAST local alignments and visually inspected the crystal structures to determine the role of the conserved lysine in substrate and protein binding or catalytic activity.
RESULTS AND DISCUSSION
Proteomic Screening of Lysine Acetylation—
Lysine acetylation is more difficult to identify by a candidate approach than protein phosphorylation due to the low radioactivity of [14C]acetyl-CoA and the weak binding affinity of anti-acetyllysine antibody. A lack of protein substrates represents one of the major bottlenecks for characterization of its biological functions. To begin the systematic study of lysine acetylation in prokaryotes, we carried out the first proteomic screening of lysine-acetylated substrates in bacteria. The goals of this study were (i) to determine the spectrum and extent of lysine acetylation in bacteria; (ii) to identify novel lysine acetylation substrates and lysine acetylation sites that could provide candidate proteins for further functional studies; and (iii) to define the molecular pathways that are likely to be affected by lysine acetylation.
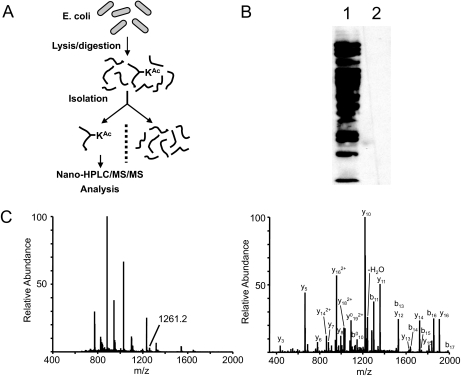
The proteomics of lysine acetylation was carried out as previously reported (11) and consisted of four steps: (i) the protein lysate of E. coli was proteolytically digested by trypsin; (ii) The resulting tryptic peptides were subjected to affinity purification by anti-acetyllysine antibody, (iii) the isolated, lysine-acetylated peptides were then analyzed by nano-HPLC/MS/MS for peptide identification and precise localization of lysine acetylation sites; and (iv) the peptide candidates were further manually evaluated to ensure the accuracy of the identification (Fig. 1, A and C). The raw spectrum of each acetylated peptide can be found in supplemental Table S1.
Fig. 1.
Synopsis of lysine acetylation proteomics. A, schematic representation of the sequential steps used for global profiling of lysine acetylation in E. coli. B, Western blotting analysis of protein lysate from E. coli with anti-acetyllysine antibody from ImmuneChem Inc. Lane 1, 30 μg of whole cell lysate from E. coli probed with anti-acetyllysine antibody; lane 2, competition with acetylated BSA (300 μg/ml). C, an example of MS and MS/MS analysis of a lysine-acetylated peptide for peptide identification and mapping of acetyllysine site in E. coli. Left panel, full MS spectrum at a retention time of 63.69 min. Right panel, MS/MS spectrum of m/z 1261.2, which identifies acetylated peptide YYQGTPSPVK*HPELTDMVIFR in isocitrate dehydrogenase, a metabolic protein in E. coli.
The strategy described here, integration of immunoisolation with mass spectrometry for characterization of biomolecules, can be traced back more than seventeen years ago (38). Peptides released from immunoisolated complexes have been analyzed by both MALDI-TOF mass spectrometry and electrospray tandem mass spectrometry for epitope mapping (39, 40), sequencing histocompatibility complex-binding peptides (41), and analysis of disease-related peptides (42). In addition to sequence-specific antibodies, pan-antibodies, such as anti-phosphotyrosine antibody, have also been used to isolate and to identify tyrosine-phosphorylated proteins on a global scale in response to extracellular stimulation (43, 44). Isolation of modified peptides from tryptic digests by immunoaffinity purification using a pan-antibody is much simpler than that of the corresponding proteins for three obvious reasons. First, the modified residue will not be buried in peptides. In contrast, the modified residue may not be accessible for antibody binding in the context of proteins due to protein-folding or non-covalent interactions (e.g. phosphotyrosine with SH2 domain). Second, a protein typically has more complex domains with variant surface properties, leading to higher non-specific binding during immunoprecipitation. Finally, a significant portion of proteins is denatured and subsequently precipitated during immunoisolation, therefore leading to more contaminant proteins. In contrast, peptides are more difficult to precipitate due to their small size and a lack of hydrophobic core structure. Nevertheless, immunoisolation using tryptic peptides has been successfully combined with mass spectrometry for proteomics of protein modifications such as tyrosine phosphorylation and lysine acetylation (11, 45).
Western blotting analysis demonstrates that proteins of a wide molecular weight range can be lysine-acetylated (Fig. 1B). Subsequent proteomics screening identified 138 lysine acetylation sites among 91 proteins (supplemental Table S2). To the best of our knowledge, none of these bacterial proteins has been associated with the modification before.
Lysine-acetylated Protein Groups in E. coli—
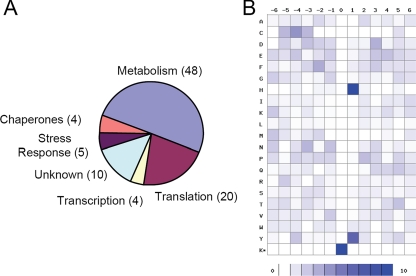
An unbiased screen of a large set of lysine acetylation substrates can provide insight into cellular pathways that would not be apparent through single-protein analysis. This has been exemplified by our recent study on proteomics of lysine acetylation in mammalian cells (11). We therefore attempted to assign each lysine acetylation substrate to a functional group based on gene ontology molecular functions or biochemical process groups, or previously published literature (Fig. 2A).
Fig. 2.
A, pie chart of functionally annotated protein groups that are lysine-acetylated. B, density map of lysine-acetylated peptides. The frequency of occurrence of amino acid residues surrounding sites of lysine acetylation was calculated, relative to the frequency of the residue within the entire E. coli genome, and schematically represented by a density map using a method described previously (11). Prevalence of specific amino acids at positions surrounding lysine acetylation sites is shown.
Several of the protein groups defined in this way are of particular interest. For example 48 of the 91 lysine acetylation substrates (∼53%) are metabolic enzymes, including 4 tricarboxylic acid (TCA) cycle proteins, 7 glycolytic enzymes, and several enzymes involved in the metabolism of nucleotides and amino acids (Table I). A few translational regulators are found to be lysine acetylation substrates in mammalian cells. However, 22% of lysine acetylation substrates identified here are proteins that are either subunits of translational machinery or are enzymes associated with translational processes, such as the aminoacyl-tRNA synthetases (supplemental Table 2). Proteins involved in stress response represented ∼5% of lysine acetylation substrates (supplemental Table 2). Transcription factors are well known examples of lysine acetylation targets in eukaryotic cells. Our screening identified two bacterial transcriptional regulators and one RNA polymerase subunit as substrates of lysine acetylation (supplemental Table 2).
Table I.
A list of metabolic proteins identified as acetylated in E. coli
Protein name, gi, general information identifier number, functional classification, and number of acetylation sites identified are indicated.
| Protein name | gi | Functional group | No. of sites |
|---|---|---|---|
| Citrate synthase | gi 16128695 | TCA cycle | 1 |
| Isocitrate dehydrogenase | gi 2618886 | TCA cycle | 1 |
| Pyruvate dehydrogenase complex, dehydrogenase component | gi 83584560 | TCA cycle | 1 |
| Dihydrolipoamide dehydrogenase | gi 26106453 | TCA cycle | 1 |
| Dihydrolipoamide acetyltransferase | gi 26246694 | TCA cycle | 1 |
| Dihydrolipoamide acyltransferase (E2) | gi 75258892 | TCA cycle | 1 |
| Succinate dehydrogenase catalytic subunit | gi 26246691 | TCA cycle | 1 |
| Phosphoglucose isomerase | gi 9664438 | Glycolysis | 2 |
| pgi | gi 68304110 | Glycolysis | 1 |
| Fructose-bisphosphate aldolase class II | gi 110643069 | Glycolysis | 1 |
| Dehydrin (fructose-bisphosphate aldolase class I) | gi 1658028 | Glycolysis | 2 |
| Enolase | gi 563868 | Glycolysis | 1 |
| Phosphoglycerate kinase | gi 26249339 | Glycolysis | 1 |
| Phosphoglyceromutase | gi 13360242 | Glycolysis | 3 |
| Glyceraldehyde-3-phosphate dehydrogenase | gi 37699654 | glycolysis | 4 |
| Pyruvate kinase | gi 26248120 | Glycolysis | 1 |
| Pyruvate kinase I | gi 147276 | Glycolysis | 2 |
| Pyruvate formate lyase subunit | gi 16130504 | Anaerobic glycolysis | 3 |
| Formate acetyltransferase 1 | gi 15800764 | Anaerobic glycolysis | 6 |
| Anaerobic class I fumarate hydratase | gi 146048 | Anaerobic glycolysis | 1 |
| Phosphoenolpyruvate carboxykinase | gi 147113 | Gluconeogenesis | 2 |
| Transketolase | gi 75227108 | Carbohydrate metabolism | 1 |
| NADP-dependent malic enzyme | gi 26109236 | Carbohydrate metabolism | 1 |
| Transketolase 2, thiamin-binding | gi 16130390 | Carbohydrate metabolism | 1 |
| Phosphopentomutase | gi 1790843 | Carbohydrate metabolism | 1 |
| NAD-dependent aldehyde dehydrogenase | gi 75515146 | Carbohydrate metabolism | 1 |
| Mannitol-1-phosphate dehydrogenase | gi 46095211 | Carbohydrate metabolism | 1 |
| 6-Phosphogluconolactonase | gi 16128735 | Carbohydrate metabolism | 1 |
| Phosphorylase, maltodextrin | gi 224195 | Carbohydrate metabolism | 1 |
| Mannose-6-phosphate isomerase | gi 1742663 | Carbohydrate metabolism | 1 |
| Fused mannose-specific PTS enzymes: IIA component/IIB component | gi 1788120 | Carbohydrate metabolism | 2 |
| Nucleoside-diphosphate-sugar epimerase | gi 75229016 | Carbohydrate metabolism | 1 |
| ADP-heptose synthase | gi 26249631 | Lipopolysaccharide biosynthesis | 1 |
| Glutamate decarboxylase isozyme | gi 15804061 | Amino acid metabolism | 3 |
| Serine hydroxymethyltransferase | gi 26248915 | Amino acid metabolism | 6 |
| Tryptophanase | gi 41936 | Amino acid metabolism | 5 |
| 3-Deoxy-d-arabino-heptulosonate-7-phosphate synthase | gi 1651339 | Amino acid metabolism | 1 |
| Glutaminase | gi 26246500 | Amino acid metabolism | 1 |
| Aminoacyl-histidine dipeptidase | gi 26246281 | Protein metabolism | 1 |
| S-Adenosylmethionine synthetase II | gi 146851 | Coenzyme metabolism | 1 |
| Purine nucleoside phosphorylase | gi 15804956 | Nucleotide salvage pathway | 1 |
| Adenylate kinase | gi 1773156 | Nucleotide metabolism | 1 |
| IMP dehydrogenase | gi 146275 | Nucleotide metabolism | 2 |
| IMP cyclohydrolase | gi 26250778 | Nucleotide metabolism | 1 |
| Ribonucleotide reductase, alpha subunit | gi 75259700 | Nucleotide metabolism | 1 |
| Deoxyribosephosphate aldolase | gi 537221 | Nucleotide metabolism | 1 |
| Glycinamide ribonucleotide transformylase | gi 16129802 | Purine biosynthesis | 1 |
| Adenylosuccinate lyase | gi 145203 | Purine biosynthesis | 2 |
Lysine-acetylated Substrate Proteins in Bacteria—
The identification of metabolic enzymes, which have previously been shown to be lysine-acetylated in the mammalian mitochondrion, represented a critical validation of functional roles of the modification in energy metabolism. Identification of a large number of metabolic enzymes and TCA proteins is reminiscent of lysine acetylation substrates identified in mitochondria, providing evidence supporting the importance of the modification in the regulation of energy metabolism.
Glycolytic Enzymes—
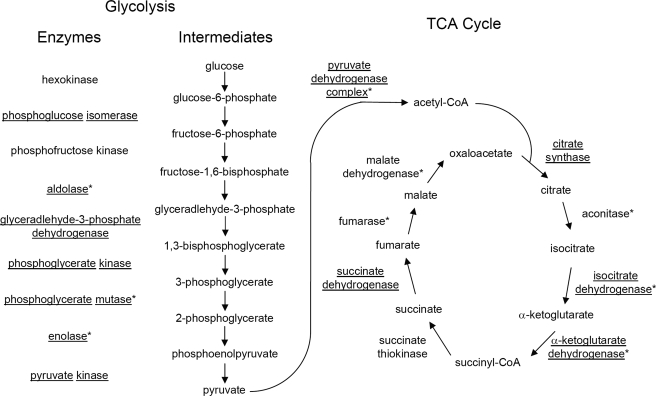
We detected 7 of 9 glycolytic enzymes, catalyzing the key reactions to degrade glucose to pyruvate, as subjects of lysine acetylation (Fig. 3). These proteins include phosphoglucose isomerase, aldolase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, phosphoglycerate mutase, enolase, and pyruvate kinase. Three of these enzymes were also found to be acetylated in mammalian samples, suggesting potentially conserved functional consequences for the modification in the regulation of glycolyic flux.
Fig. 3.
Lysine-acetylated proteins involved in glucose degradation and TCA cycle. Proteins identified as lysine-acetylated in the E. coli screen are underlined. Those identified in a screen of mammalian cells (11) are marked with an asterisk.
Pyruvate Dehydrogenases—
Pyruvate is one of the major products of glycolysis. It can be cleaved by either pyruvate dehydrogenase or pyruvate formate lyase, of which both were found to be lysine-acetylated. The oxidative decarboxylation of pyruvate by pyruvate dehydrogenase generates acetyl-CoA and one NADH and is therefore functionally restricted to respiratory metabolism under aerobic conditions. All three subunits of pyruvate dehydrogenase are subjects of lysine acetylation. On the other hand, non-oxidative cleavage of pyruvate to acetyl-CoA and formate is catalyzed by pyruvate formate lyase, and this enzyme is functional only anaerobically. The activities of the two enzymes are usually mutually exclusive. Pyruvate formate lyase is a homodimer that catalyzes the conversion of pyruvate to acetyl-CoA, which is in turn used to produce ATP through acetyl phosphate. The ATP produced in this manner is the single ATP source in E. coli cells under anaerobic conditions with pyruvate as the sole carbon and energy source (46). In E. coli cells, the activity of the protein is tightly regulated at both the transcriptional level and by post-translational modification.
TCA Cycle Proteins—
Four of the eight TCA cycle proteins, citrate synthase, isocitrate dehydrogenase, and succinate dehydrogenase catalytic subunits, 2-ketoglutarate dehydrogenase (E3 dihydrolipoamide dehydrogenase subunit shared by both 2-ketoglutarate dehydrogenase and pyruvate dehydrogenase) are lysine-acetylated (Fig. 3). Among the four proteins, citrate synthase and isocitrate dehydrogenase are known to be regulated at the level of transcription and protein modification. The synthesis of citrate synthase is subject to catabolite repression. It is repressed by glucose and anaerobiosis and induced by acetate and oxygen. Isocitrate dehydrogenase catalyzes the conversion of isocitrate to α-ketoglutarate and CO2, which can be a rate-limiting step in the citric acid cycle, and is also involved in the regulation of carbon flux at the branch point between the TCA and glyoxylate cycles. Protein phophorylation is also known to regulate the protein functions (47, 48).
Enzymes involved in the metabolism of amino acids and nucleotides are also lysine-acetylated. These proteins include 7 proteins involved in amino acid metabolism and 7 proteins in nucleotide metabolism.
One enzyme involved in nucleotide metabolism, deoxyribosephosphate aldolase, is of particular interest. The enzyme catalyzes the reversible reaction between glyceraldehyde-3-phosphate and acetaldehyde to form 2-deoxyribose-5-phosphate. We identified this enzyme as being acetylated at lysine 167. This residue has been implicated in the catalytic mechanism of the enzyme, forming a Schiff base with the substrate (49). Acetylation of this active site lysine would therefore be expected to block enzymatic activity. Such acetylation of active site lysine may represent a common regulatory mechanism among the aldolase family of proteins. Our previous survey of lysine acetylation among mammalian proteins identified acetylation of lysine 146 of the glycolytic enzyme fructose-1, 6-bisphosphate aldolase (11), which is also the key active site catalytic residue involved in Schiff base formation.
Transcription and Translation Factors—
Transcription factors and histones are founding members of lysine acetylation substrates in mammalian cells. Reminiscent of this, two transcription factors (cAMP receptor protein and trp repressor-binding protein) and one subunit of RNA polymerase (rpoB) are lysine acetylation substrates. In addition to transcriptional regulators, 20 proteins involved in translational regulation are subjects of lysine acetylation. These proteins include translational elongation factors, ribosomal proteins as well as aminoacyl-tRNA synthetases. Lysine acetylation has not been detected in ribosomal proteins or in aminoacyl-tRNA synthetases in mammals, suggesting that the modification of such protein may have been lost during evolution.
A large portion of cellular energy is used in protein synthesis. Therefore, translational machinery needs to be synchronized with energy availability. It is not surprising that the expression level of the protein for about half of the aminoacyl-tRNA synthetases and for translational factors are under metabolic control (50). Our screening identified lysine acetylation at 5 aminoacyl-tRNA synthetases, 11 ribosomal subunits, and three translational machinery proteins, implying that post-translational modifications might provide an alternative avenue to modulate the activity of cellular translational machinery.
Stress Response Proteins—
In eukaryotic cells, activation of the sirtuin family of deacetylases promotes cell survival and resistance to stress, implying that the lysine acetylation status of proteins may be altered in response to stress. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan (51). Our previous screening identified heat shock proteins as well as reactive oxygen species regulators such as superoxide dismutase as substrates of lysine acetylation (11). Like eukaryotic cells, a significant portion of stress response proteins in E. coli are also substrates of lysine acetylation, including chaperones and proteins involved in regulation of free radical reduction (such as superoxide dismutase, alkyl hydroperoxide reductase, and thioredoxin). In E. coli, loss of alkyl hydroperoxide reductase and SOD leads to an increased sensitivity to hydrogen peroxide and alkyl hydroperoxide oxidative stress (52, 53).
Several respiratory components of the mammalian mitochondrion are closely related to those in E. coli. Mitochondrial NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), and cytochrome c oxidase (complex IV) are mammalian homologs of E. coli proteins that are lysine-acetylated.
Evolutionary Conservation of Lysine Acetylation between E. coli and Mammalian Cells—
The mitochondrion is considered to have evolved from α-proteobacteria. Phylogenetic studies suggest that about one tenth of yeast mitochondrial proteins could be evolved from ancestral free-living α-proteobacteria whereas the remaining 90% are recruited from the nuclear genome of the eukaryotic host (54).
The results of our screen for lysine-acetylated proteins in E. coli suggest that regulatory programs tuning the activities of metabolic enzymes may be conserved between E. coli and mammalian cells. This concept is supported by the overlap in protein identities between the mammalian and E. coli datasets and the evidence of a common regulatory modification observed among the active site lysine of aldolase family members. Twenty-two of the 91 E. coli lysine-acetylated proteins (∼25%) have mammalian homologs, based on BLAST sequence alignment with 25% sequence identity as the cutoff score (supplemental Table 2). We believe that the number is significantly underestimated because of the low percentage of lysine-acetylated proteins identified in mammalian cells that was caused by the wide dynamic range and limited sensitivity in our previous proteomics screening (11). Identification of a large number of lysine-acetylated orthologs between E. coli and mammalian cells, the abundance of lysine acetylation in both mitochondrion and E. coli, and the evolutionary linkage between the mitochondrion and E. coli suggest that the modification is likely to be evolutionarily conserved from bacteria to mammalian cells.
Lysine Acetylation Motifs in Bacterial Substrate Proteins and Bioinformatics Analysis—
Conserved protein sequence motifs are associated with some post-translational modifications such as protein phosphorylation. Structural studies of GCN5 HAT and H3 tail peptides suggest a recognition site of GKXP (55). The motifs for lysine acetylation remain largely unknown. The dataset of acetylation sites identified from E. coli proteins allows us to conduct a preliminary analysis of motif preference (Fig. 2B). Such analysis identified histidine and tyrosine as preferred amino acid residues at the +1 position.
In contrast to the E. coli lysine acetylation motifs, mammalian lysine acetylation substrates in general show less preference for residues surrounding acetyllysine residues (11). This could be caused by averaged motif information from diverse acetyltransferases. Alternatively, substrates may be recognized through the three-dimensional structure of the substrate proteins rather than their linear sequence. However, when considering the mitochondrial subset of lysine acetylation substrates identified in the mammalian dataset, a preference for histidine and tyrosine at the +1 position was also observed (11). The preference of amino acid residues flanking acetyllysine residues in E. coli proteins suggests that the modification might be catalyzed by a limited subset of acetyltransferases with unique substrate preferences. In addition, related acetyltransferases may be at work in the case of mitochondrial acetylation. Our results suggest the possibility that linear lysine acetylation motifs exist among lysine-acetylated proteins similar to motifs observed for phosphorylation.
To evaluate the structural features of acetyllysines in the substrate proteins, we first identified those proteins for which a homologous crystal structure was available. BLAST local alignment was used to map the position of the acetylated lysine onto the crystal structure. Visual inspection of the available crystal structures using the Pymol program suggested that most of the acetylation sites (95%) identified occur on the surface of proteins, rather than at active site or ligand binding residues. However, in some instances, modification sites were found within oligomerization interfaces, such as in serine hydroxymethyltransferase (Protein Data Bank code 1dfo) (56), isocitrate dehydrogenase (Protein Data Bank code 1hqs) (57), and IMP cyclohydrolase (Protein Data Bank code 1pkx) (58). This suggests that acetylation may affect the oligomerization of proteins or the formation of protein complexes. Such effects of lysine acetylation are known to occur in eukaryotes through the specific binding of acetylated lysine by bromodomains.
Lysine Acetylation Is a Regulatory Post-translational Modification in E. coli—
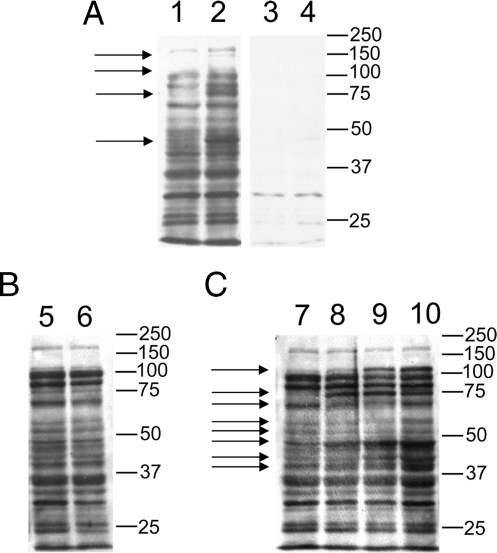
To test if lysine acetylation profiles can be regulated in E. coli, we carried out Western blotting analysis. The protein lysates from a wild-type E. coli strain (MG1655) and a CobB-deficient strain (JW1106) were prepared and resolved in SDS-PAGE to detect lysine-acetylated proteins by Western blotting analysis using an anti-acetyllysine antibody (Fig. 4A). Competition experiments using acetylated BSA suggest that most of the signals detected are specific to acetyllysine (Fig. 4A, lanes 1 and 2 versus lanes 3 and 4). Interestingly, the signals for four major bands increased between the wild type and CobB-deficient strain, whereas the majority was unchanged (Fig. 4A), suggesting the existence of other deacetylases in addition to CobB.
Fig. 4.
Western blotting analysis of protein lysate with an anti-acetyllysine antibody from ImmuneChem Inc. The 40-μg protein lysate was resolved in 4–20% SDS-PAGE. The Western blotting analysis was carried out as described previously (11). Each of the gel bands that have significant differences between the two E. coli strains or among the four oxygen conditions is labeled with an arrow on the left. The molecular weight markers are labeled on the right. A, lysine acetylation profiles of two E. coli strains (Lanes 1 and 3, MG1655; Lanes 2 and 4, JW1106). Lanes 1 and 2, probed with anti-acetyllysine antibody and non-acetylated BSA (300 μg/2 ml); Lanes 3 and 4, probed with anti-acetyllysine antibody and acetylated BSA (300 μg/2 ml). B, impact of nutritional conditions (E. coli MG1655). Lane 5, control and Lane 6, starvation. C, influences of oxygen conditions (E. coli MG1655). Lane 7, control (100% air); Lane 8, 25% air; Lane 9, 5% air; and Lane 10, 100% nitrogen.
To further demonstrate that lysine acetylation profiles may be altered in response to stress, lysine acetylation profiles were analyzed by Western blotting analysis using the E. coli cell lysates from cells that were either starved or subjected to hypoxic conditions. Starvation has no obvious effect on the lysine acetylation profile in E. coli (Fig. 4B). However, when E. coli was exposed to hypoxia, significant changes were observed (Fig. 4C), suggesting that lysine acetylation is a dynamic and regulated process in E. coli.
It should be noted that our approach is capable of detecting only the most abundant substrate proteins. It is possible that changes in lysine acetylation for other less abundant proteins and substrate proteins with sequence motifs not recognized by the antibodies escaped our detection.
Several important points have emerged from this comprehensive protein screening of protein lysine acetylation. First, a large number of previously unknown lysine acetylation substrates exist in prokaryotic species. Given the key roles of many of these substrate proteins in cellular functions and high sequence conservation among prokaryotic species, it is highly likely that the modification is conserved and shares similar functions among prokaryotic species other than E. coli. Second, two preferred residues, histidine and tyrosine, at the +1 position of acetyllysine were identified, implying that linear sequence might be important for substrate binding and/or activity of acetyltransferase(s) in bacteria. This finding is intriguing given that a similar preference was observed among lysine-acetylated substrates identified in a screen of mitochondrial proteins, suggesting the existence of a related set of acetyltransferases in mitochondria (11). Third, the spectrum of lysine acetylation substrates is evolutionarily well conserved. A comparison of lysine acetylation substrates from E. coli and mammalian cell lines suggests that three groups of substrate proteins are enriched in lysine acetylation, including metabolic enzymes, transcriptional/translational regulators, and stress response proteins. Identification of a high number of ribosomal proteins and aminoacyl-tRNA synthetases was a surprise to us as these proteins were abundant in eukaryotic cells, and lysine acetylation was not detected among these proteins in mammalian cells. Lysine acetylation of these proteins might have been lost during evolution.
Like our initial studies of lysine acetylation in mammalian cells (11), our study has limitations. First, only protein substrates present in medium to high abundance were identified. We might miss a significant portion of lysine acetylation substrates that are either of low abundance or modified at a low stoichiometry. Second, dynamic analysis has not been carried out under diverse genetic backgrounds and nutrient sources. Third, substrates of known acetyltransferases and deacetylases have not been defined. The answers for these questions await future research by quantitative proteomics of lysine acetylation with high sensitivity (e.g. protein pre-fractionation before proteomics studies).
Identification of a large number of lysine acetylation substrate raises many interesting questions. Only one acetyltransferase and one deacetylase have been described in prokaryotes. Identification of a large number of lysine acetylation substrate proteins raises the possibility of additional enzymes in bacteria that regulate the modification status. The recent discovery of a novel yeast acetyltransferase with no apparent sequence similarity with existing ones further supports such a possibility (59). Recently, we identified two novel in vivo lysine modifications, lysine propionylation at Lys-5, Lys-8, and Lys-12 and lysine butyrylation at Lys-5 and Lys12 of histone H4 (25). These lysine residues were found to be acetylated and methylated before. Interestingly, two acetyltransferases, p300/CBP, can carry out in vitro lysine propionylation and lysine butyrylation in vitro on histones H3 and H4. It would be intriguing to know if the lysine-acetylated substrates can also be lysine-propionylated or lysine-butyrylated and the identities of the regulatory enzymes.
Our results provide a large number of future research opportunities in prokaryotic biology. The large datasets will provide protein leads for further genetic and molecular biological studies to test roles of lysine acetylation sites in cellular physiology. Prokaryotic species, such as E. coli, have been genetically engineered to boost protein expression or for fermentation of special chemicals, such as industrial ethanol and glycerol. Such genetic engineering typically takes advantage of overexpression of specific proteins and/or improving the activities of an enzyme with an optimized sequence. Nevertheless, these genetically engineered species have not fully taken advantage of the regulatory lysine acetylation pathway. Given the essential roles of lysine acetylation in diverse metabolic pathways and importance of acetyl Co-A and NAD (the co-factors for lysine acetylation regulatory enzymes) in cellular metabolism, the modification is likely to play an important role in bacterial physiology. Therefore, understanding, capturing, and optimizing the modification landscape might provide a novel approach for the engineering of industrial bacterial species with high efficiency. The bioterror threat posed from certain bacteria (such as Bacillus anthracis) and the emergence of drug-resistant bacteria remind us that the biology of prokaryotes remains to be further studied. Unfortunately, post-translational modifications in prokaryotes have been overlooked in the past. Generation of lysine acetylation datasets reminds us of the need for further studies of modification pathways in prokaryotes, of which some might provide good avenues for therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Marie-Alda Gilles-Gonzalez and Zhihong Zhang for helpful suggestions. We are grateful to Xiang-Jiao Yang at McGill University for critical reading of the manuscript and helpful comments.
Footnotes
Published, MCP Papers in Press, August 23, 2008, DOI 10.1074/mcp.M800187-MCP200
The abbreviations used are: BSA, bovine serum albumin; MS, mass spectroscopy; CoA, coenzyme A; HPLC, high performance liquid chromatography; LC/MS/MS, liquid chromatography tandem mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grant CA107943 (to Y. Z.). This work was also supported by the Robert A. Welch Foundation Grant I-1550 (to Y. Z.), and National Technology Centers for Networks and Pathways Grant U45 RR020839 (to H. Z.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcp.org) contains Supplemental Tables S1 and S2. The raw MS/MS spectra of all the acetylated peptides and the corresponding acetylated proteins identified in this screen are available as supplemental information.
REFERENCES
- 1.Haigis, M. C., and Guarente, L. P. ( 2006) Mammalian sirtuins-emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20, 2913–2921 [DOI] [PubMed] [Google Scholar]
- 2.Hake, S. B., Xiao, A., and Allis, C. D. ( 2004) Linking the epigenetic “language” of covalent histone modifications to cancer. Br. J. Cancer 90, 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinsey, T. A., and Olson, E. N. ( 2004) Cardiac histone acetylation – therapeutic opportunities abound. Trends Genet. 20, 206–213 [DOI] [PubMed] [Google Scholar]
- 4.Yang, X. J. ( 2004) The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32, 959–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouzarides, T. ( 2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19, 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarente, L., and Picard, F. ( 2005) Calorie restriction – the SIR2 connection. Cell 120, 473–482 [DOI] [PubMed] [Google Scholar]
- 7.Cohen, H. Y., Miller, C., Bitterman, K. J., Wall, N. R., Hekking, B., Kessler, B., Howitz, K. T., Gorospe, M., de Cabo, R., and Sinclair, D. A. ( 2004) Calorie restriction promotes mammalian cell survival by inducing SIRT1 deacetylase. Science 305, 390–393 [DOI] [PubMed] [Google Scholar]
- 8.Cohen, T., and Yao, T. P. ( 2004) AcK-knowledge reversible acetylation. Sci. STKE 2004, pe42. [DOI] [PubMed] [Google Scholar]
- 9.Michishita, E., Park, J. Y., Burneskis, J. M., Barrett, J. C., and Horikawa, I. ( 2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baur, J. A., Pearson, K. J., Price, N. L., Jamieson, H. A., Lerin, C., Kalra, A., Prabhu, V. V., Allard, J. S., Lopez-Lluch, G., Lewis, K., Pistell, P. J., Poosala, S., Becker, K. G., Boss, O., Gwinn, D., Wang, M., Ramaswamy, S., Fishbein, K. W., Spencer, R. G., Lakatta, E. G., Le Couteur, D., Shaw, R. J., Navas, P., Puigserver, P., Ingram, D. K., de Cabo, R., and Sinclair, D. A. ( 2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, S. C., Sprung, R., Chen, Y., Xu, Y., Ball, H., Pei, J., Cheng, T., Kho, Y., Xiao, H., Xiao, L., Grishin, N. V., White, M., Yang, X. J., and Zhao, Y. ( 2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 12.Schwer, B., Bunkenborg, J., Verdin, R. O., Andersen, J. S., and Verdin, E. ( 2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U. S. A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallows, W. C., Lee, S., and Denu, J. M. ( 2006) Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U. S. A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lombard, D. B., Alt, F. W., Cheng, H. L., Bunkenborg, J., Streeper, R. S., Mostoslavsky, R., Kim, J., Yancopoulos, G., Valenzuela, D., Murphy, A., Yang, Y., Chen, Y., Hirschey, M. D., Bronson, R. T., Haigis, M., Guarente, L. P., Farese, R. V., Jr., Weissman, S., Verdin, E., and Schwer, B. ( 2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pesavento, J. J., Kim, Y. B., Taylor, G. K., and Kelleher, N. L. ( 2004) Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J. Am. Chem. Soc. 126, 3386–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyne, M. T., II, Pesavento, J. J., Mizzen, C. A., and Kelleher, N. L. ( 2006) Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J. Proteome Res. 5, 248–253 [DOI] [PubMed] [Google Scholar]
- 17.Medzihradszky, K. F., Zhang, X., Chalkley, R. J., Guan, S., McFarland, M. A., Chalmers, M. J., Marshall, A. G., Diaz, R. L., Allis, C. D., and Burlingame, A. L. ( 2004) Characterization of Tetrahymena histone H2B variants and post-translational populations by electron capture dissociation (ECD) fourier transform ion cyclotron mass spectrometry (FT-ICR MS). Mol. Cell. Proteomics 3, 872–886 [DOI] [PubMed] [Google Scholar]
- 18.Hake, S. B., Garcia, B. A., Duncan, E. M., Kauer, M., Dellaire, G., Shabanowitz, J., Bazett-Jones, D. P., Allis, C. D., and Hunt, D. F. ( 2006) Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 281, 559–568 [DOI] [PubMed] [Google Scholar]
- 19.Beck, H. C., Nielsen, E. C., Matthiesen, R., Jensen, L. H., Sehested, M., Finn, P., Grauslund, M., Hansen, A. M., and Jensen, O. N. ( 2006) Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteomics 5, 1314–1325 [DOI] [PubMed] [Google Scholar]
- 20.Gray, M. W., Burger, G., and Lang, B. F. ( 1999) Mitochondrial evolution. Science 283, 1476–1481 [DOI] [PubMed] [Google Scholar]
- 21.Starai, V. J., Celic, I., Cole, R. N., Boeke, J. D., and Escalante-Semerena, J. C. ( 2002) Sir2-Dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390–2392 [DOI] [PubMed] [Google Scholar]
- 22.Barak, R., and Eisenbach, M. ( 2001) Acetylation of the response regulator, CheY, is involved in bacterial chemotaxis. Mol. Microbiol. 40, 731–743 [DOI] [PubMed] [Google Scholar]
- 23.Bell, S. D., Botting, C. H., Wardieworth, B. N., Jackson, S. P., and White, M. F. ( 2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296, 148–151 [DOI] [PubMed] [Google Scholar]
- 24.Zhao, K., Chai, X., and Marmorstein, R. ( 2003) Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA binding. J. Biol. Chem. 278, 26071–26077 [DOI] [PubMed] [Google Scholar]
- 25.Chen, Y., Sprung, R., Tang, Y., Ball, H., Sangras, B., Kim, S. C., Falck, J. R., Peng, J., Gu, W., and Zhao, Y. ( 2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blander, G., and Guarente, L. ( 2004) The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 27.Starai, V. J., and Escalante-Semerena, J. C. ( 2004) Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 28.Starai, V. J., Takahashi, H., Boeke, J. D., and Escalante-Semerena, J. C. ( 2003) Short-chain fatty acid activation by acetyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K. A., Tomita, M., Wanner, B. L., and Mori, H. ( 2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiang, L., Xiao, H., Campos, E. I., Ho, V. C., and Li, G. ( 2005) Development of a PAN-specific, affinity-purified anti-acetylated lysine antibody for detection, identification, isolation, and intracellular localization of acetylated protein. J. Immunoassay Immunochem. 26, 13–23 [DOI] [PubMed] [Google Scholar]
- 31.Zhang, H., Zha, X., Tan, Y., Hornbeck, P. V., Mastrangelo, A. J., Alessi, D. R., Polakiewicz, R. D., and Comb, M. J. ( 2002) Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J. Biol. Chem. 277, 39379–39387 [DOI] [PubMed] [Google Scholar]
- 32.Kim, S. C., Chen, Y., Mirza, S., Xu, Y., Lee, J., Liu, P., and Zhao, Y. ( 2006) A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J. Proteome Res. 5, 3446–3452 [DOI] [PubMed] [Google Scholar]
- 33.Chen, Y., Kwon, S. W., Kim, S. C., and Zhao, Y. ( 2005) Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J Proteome Res. 4, 998–1005 [DOI] [PubMed] [Google Scholar]
- 34.Weiss, B. ( 2006) Evidence for mutagenesis by nitric oxide during nitrate metabolism in Escherichia coli. J. Bacteriol. 188, 829–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos, J. M., Freire, P., Vicente, M., and Arraiano, C. M. ( 1999) The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol. Microbiol. 32, 789–798 [DOI] [PubMed] [Google Scholar]
- 36.Chandonia, J. M., Hon, G., Walker, N. S., Lo Conte, L., Koehl, P., Levitt, M., and Brenner, S. E. ( 2004) The ASTRAL compendium in 2004. Nucleic Acids Res. 32, D189–D192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murzin, A. G., Brenner, S. E., Hubbard, T., and Chothia, C. ( 1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540 [DOI] [PubMed] [Google Scholar]
- 38.Suckau, D., Kohl, J., Karwath, G., Schneider, K., Casaretto, M., Bitter-Suermann, D., and Przybylski, M. ( 1990) Molecular epitope identification by limited proteolysis of an immobilized antigen-antibody complex and mass spectrometric peptide mapping. Proc. Natl. Acad. Sci. U. S. A. 87, 9848–9852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, Y., and Chait, B. T. ( 1994) Protein epitope mapping by mass spectrometry. Anal. Chem. 66, 3723–3726 [DOI] [PubMed] [Google Scholar]
- 40.Papac, D. I., Hoyes, J., and Tomer, K. B. ( 1994) Epitope mapping of the gastrin-releasing peptide/anti-bombesin monoclonal antibody complex by proteolysis followed by matrix-assisted laser desorption ionization mass spectrometry. Protein Sci. 3, 1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunt, D. F., Henderson, R. A., Shabanowitz, J., Sakaguchi, K., Michel, H., Sevilir, N., Cox, A. L., Appella, E., and Engelhard, V. H. ( 1992) Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 [DOI] [PubMed] [Google Scholar]
- 42.Yip, T. T., Van de Water, J., Gershwin, M. E., Coppel, R. L., and Hutchens, T. W. ( 1996) Cryptic antigenic determinants on the extracellular pyruvate dehydrogenase complex/mimeotope found in primary biliary cirrhosis. A probe by affinity mass spectrometry. J. Biol. Chem. 271, 32825–32833 [DOI] [PubMed] [Google Scholar]
- 43.Pandey, A., Podtelejnikov, A. V., Blagoev, B., Bustelo, X. R., Mann, M., and Lodish, H. F. ( 2000) Analysis of receptor signaling pathways by mass spectrometry: identification of Vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc. Natl. Acad. Sci. U. S. A. 97, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandey, A., Fernandez, M. M., Steen, H., Blagoev, B., Nielsen, M. M., Roche, S., Mann, M., and Lodish, H. F. ( 2000) Identification of a novel immunoreceptor tyrosine-based activation motif-containing molecule, STAM2, by mass spectrometry and its involvement in growth factor and cytokine receptor signaling pathways. J. Biol. Chem. 275, 38633–38639 [DOI] [PubMed] [Google Scholar]
- 45.Rush, J., Moritz, A., Lee, K. A., Guo, A., Goss, V. L., Spek, E. J., Zhang, H., Zha, X. M., Polakiewicz, R. D., and Comb, M. J. ( 2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 46.Pascal, M. C., Chippaux, M., Abou-Jaoude, A., Blaschkowski, H. P., and Knappe, J. ( 1981) Mutants of Escherichia coli Lys-12 with defects in anaerobic pyruvate metabolism. J. Gen. Microbiol. 124, 35–42 [DOI] [PubMed] [Google Scholar]
- 47.Borthwick, A. C., Holms, W. H., and Nimmo, H. G. ( 1984) Amino acid sequence round the site of phosphorylation in isocitrate dehydrogenase from Escherichia coli ML308. FEBS Lett. 174, 112–115 [DOI] [PubMed] [Google Scholar]
- 48.Thorsness, P. E., and Koshland, D. E., Jr. ( 1987) Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J. Biol. Chem. 262, 10422–10425 [PubMed] [Google Scholar]
- 49.Heine, A., Luz, J. G., Wong, C. H., and Wilson, I. A. ( 2004) Analysis of the class I aldolase binding site architecture based on the crystal structure of 2-deoxyribose-5-phosphate aldolase at 0.99 Å resolution. J. Mol. Biol. 343, 1019–1034 [DOI] [PubMed] [Google Scholar]
- 50.Neidhardt, F. (ed) ( 1996) Escherichia coli and Salmonella Cellular and Molecular Biology, American Society for Microbiology, Washington DC
- 51.Howitz, K. T., Bitterman, K. J., Cohen, H. Y., Lamming, D. W., Lavu, S., Wood, J. G., Zipkin, R. E., Chung, P., Kisielewski, A., Zhang, L. L., Scherer, B., and Sinclair, D. A. ( 2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196 [DOI] [PubMed] [Google Scholar]
- 52.Imlay, J. A., and Linn, S. ( 1987) Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J. Bacteriol. 169, 2967–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beyer, W., Imlay, J., and Fridovich, I. ( 1991) Superoxide dismutases. Prog. Nucleic Acids Res. Mol. Biol. 40, 221–253 [DOI] [PubMed] [Google Scholar]
- 54.Karlberg, O., Canback, B., Kurland, C. G., and Andersson, S. G. ( 2000) The dual origin of the yeast mitochondrial proteome. Yeast 17, 170–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rojas, J. R., Trievel, R. C., Zhou, J., Mo, Y., Li, X., Berger, S. L., Allis, C. D., and Marmorstein, R. ( 1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401, 93–98 [DOI] [PubMed] [Google Scholar]
- 56.Scarsdale, J. N., Radaev, S., Kazanina, G., Schirch, V., and Wright, H. T. ( 2000) Crystal structure at 2.4 A resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate. J. Mol. Biol. 296, 155–168 [DOI] [PubMed] [Google Scholar]
- 57.Singh, S. K., Matsuno, K., LaPorte, D. C., and Banaszak, L. J. ( 2001) Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 A. Insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase. J. Biol. Chem. 276, 26154–26163 [DOI] [PubMed] [Google Scholar]
- 58.Wolan, D. W., Cheong, C. G., Greasley, S. E., and Wilson, I. A. ( 2004) Structural insights into the human and avian IMP cyclohydrolase mechanism via crystal structures with the bound XMP inhibitor. Biochemistry 43, 1171–1183 [DOI] [PubMed] [Google Scholar]
- 59.Driscoll, R., Hudson, A., and Jackson, S. P. ( 2007) Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315, 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.