Abstract
The 20 S proteasome complexes are major contributors to the intracellular protein degradation machinery in mammalian cells. Systematic administration of proteasome inhibitors to combat disease (e.g. cancer) has resulted in positive outcomes as well as adversary effects. The latter was attributed to, at least in part, a lack of understanding in the organ-specific responses to inhibitors and the potential diversity of proteomes of these complexes in different tissues. Accordingly, we conducted a proteomic study to characterize the 20 S proteasome complexes and their postulated organ-specific responses in the heart and liver. The cardiac and hepatic 20 S proteasomes were isolated from the same mouse strain with identical genetic background. We examined the molecular composition, complex assembly, post-translational modifications and associating partners of these proteasome complexes. Our results revealed an organ-specific molecular organization of the 20 S proteasomes with distinguished patterns of post-translational modifications as well as unique complex assembly characteristics. Furthermore, the proteome diversities are concomitant with a functional heterogeneity of the proteolytic patterns exhibited by these two organs. In particular, the heart and liver displayed distinct activity profiles to two proteasome inhibitors, epoxomicin and Z-Pro-Nle-Asp-H. Finally, the heart and liver demonstrated contrasting regulatory mechanisms from the associating partners of these proteasomes. The functional heterogeneity of the mammalian 20 S proteasome complexes underscores the concept of divergent proteomes among organs in the context of an identical genome.
The proteasomes are enzymatic multi-protein complexes that are central to the ubiquitin-proteasome system. Proteasome complexes are found in diverse organisms and exist in all mammalian cell types. Multiple investigations document a defective ubiquitin-proteasome system in many human diseases. The reported pathogeneses are diverse, and the disease phenotypes are steadily increasing, with most investigative efforts being focused on the involvement of proteasomes in cancer. Inhibition of proteasomes has been found to be significantly beneficial for treating multiple myelomas and other forms of oncogenesis (1). However, recent reports documented dramatic side effects of proteasome inhibitors on other organs, particularly the heart (2–4), whereas the underlying mechanism is not understood. We postulate that tissue heterogeneity of proteasome structure and function exists, which may contribute to the dichotomous responses observed in different organs.
The proteolytic activities of the proteasome emanate from the core enzymes of the 20 S complexes. The 20 S proteasome is important for degrading oxidized proteins and has also been shown to degrade non-oxidized and non-ubiquitinated substrates, such as ornithine decarboxylase, p53 and p73 (5, 6). Every 20 S proteasome is composed of four stacked rings, with the inner rings containing seven β subunits (forming the central catalytic chamber) and the outer rings containing seven α subunits. Three of the β subunits (β1, β2, β5) are post-translationally cleaved at their amino terminus yielding active proteases (7). These three proteolytically active β subunits (β1, β2, β5) can be replaced with inducible counterparts (β1i, β2i, β5i) (8). The introduction of inducible subunits into 20 S proteasomes provokes a change in complex assembly, altering their proteolytic substrate specificity. A variable molecular organization of the 20 S complexes provides the cell with a dynamic range of proteolytic capacities and affords the potential for functional heterogeneties (9–11).
In this investigation, we examined the molecular composition, complex assembly, and post-translational modifications of the cardiac and the hepatic 20 S proteasomes from the same animal strain. Furthermore, we evaluated the functional impact of the diverse 20 S proteome biology in two different organs. Using blue-native polyacrylamide gel electrophoresis (BN-PAGE)1 and subsequent LC-MS/MS analyses, we delineated the molecular organization of the native 20 S proteasome complexes and their associating partners. This is the first proteomic report regarding organ-specific responses to proteasome inhibition. Our data demonstrated significant heterogeneity in the proteome biology and proteolytic function of the 20 S proteasome complexes in these organs.
MATERIALS AND METHODS
20 S Proteasome Purification—
The 20 S proteasome complexes were purified from the heart and liver of ICR mice, using a previously described method (10). Briefly 10 g of tissue (heart or liver) was homogenized by a polytron homogenizer in homogenizing buffer (20 mm Tris-HCl, pH 7.8, 0.1 mm EDTA, 1 mm DTT, protease inhibitor mixture from Roche, and phosphatase inhibitor mixture from Sigma). The homogenate was centrifuged for 2 h at 25,000 × g to remove the nuclear and mitochondrial fractions. The resulting supernatant (cytosolic fraction) was then precipitated with ammonium sulfate and the pellet between 40% to 60% ammonium sulfate saturation was collected and resuspended in 10 ml of dialysis buffer (20 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 0.5 mm DTT). The pellet was dialyzed against 4 liters of dialysis buffer overnight). The dialysate was then separated on a strong anion exchange column (Q FastFlow XK 26/40 from GE Healthcare) by stepwise salt elution (200 ml of 45% buffer B; then 200 ml of 75% buffer B, and finally 200 ml of 100% buffer B. Buffer A: 20 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 0.5 mm DTT, glycerol 10%; buffer B: 20 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 0.5 mm DTT, glycerol 10%, 600 mm KCl). Enriched 20 S proteasomes were recovered from the 75% buffer B elution peak and centrifuged for 19 h at 42,000 rpm (4 °C; Ti 45 fixed angle rotor from Beckman). The pellet was collected and further resolved by another strong ion-exchange column (MonoQ GL5/50 from GE Healthcare) by gradient elution of 17.5 column volumes. The purified 20 S proteasomes were recovered at ∼60% buffer B.
BN-PAGE—
For separation of the purified 20 S proteasomes by BN-PAGE, 1–3 μg of purified proteasome sample (at a concentration of 1 μg/μl) was mixed with BN sample buffer (15 μl). BN-gels were prepared as described previously (12). Briefly, 6% gels were cast in a Bio-Rad Protean II minigel system. Ferritin (440 and 880 kDa; Sigma-Aldrich) was used as the molecular weight marker. The buffers were 50 mm bis-Tris, pH 7.0 for the anode and 50 mm tricine, 15 mm bis-Tris, 0.02% Coomassie G250 for the cathode. Protein samples were adjusted with a concentrated BN sample buffer to a final concentration of 75 mm aminohexanoic acid, 5 mm bis-Tris, and 0.05% Coomassie G250. For sample migration through the stacking and the resolving gel, voltages were set to 100 and 500 V, respectively. After one-third of the run time, the Coomassie-stained cathode buffer was replaced with buffer containing 0.002% Coomassie G250.
Antibodies, Immunoblot Analysis, and Confocal Immunohistochemistry—
Immune complexes were detected using an Odyssey system (LI-COR). Purified 20 S proteasomes were investigated by immunoblotting using 20 S antibodies (α3 (PW8115), α7 (PW8110), β1 (PW8140), β2 (PW9300), β5 (PW8895), β1i (PW8345), β2i (PW8350), β5i (PW8200), anti-polyubiquitin (UG9510), anti-Sumo-1 (PW9460), anti-core (PW8155), anti-Rpt4 (PW8830), and anti-NEED8 (PW9340) from Biomol) and anti-methylarginine (7E6, Abcam), anti-methyllysine (ab23366, Abcam), anti-glutathione (MAB5310; Chemicon) and acetyl lysine (05–515) from Upstate. Phosphorylated proteins were detected using an antibody mixture from Calbiochem, providing a more complete profile of phosphoamino acid epitopes: anti-phosphoserine antibodies 1C8, 4A3, 4A9, and 16B4 were combined and used to detect phosphorylated serine residues, whereas anti-phosphothreonine antibodies 1E11, 4D11, and 14B3 and anti-phosphotyrosine antibodies 3B12, 2C8, 16F4, and 9H8 were combined and used for phosphorylated threonine or tyrosine residues, respectively. Protein loading was controlled by Ponceau S staining of transblots before immunoblotting and is representative of at least three replicates (n = 3).
Freshly isolated adult cardiomyocytes were isolated as described previously (13, 14) and were plated on polylysine precoated coverslips (1 h at room temperature). The cells were then fixed in cold acetone (10 min at −20 °C) or in 4% paraformaldehyde (20 min at room temperature). Nonspecific binding was blocked using 10% goat or donkey serum, 0.2% Triton X-100 in phosphate-buffered saline (PBS), pH 7.4 for 30 min at room temperature. The cells were incubated overnight at 4 °C with the primary antibody in 1% goat or donkey serum, 0.2% Triton X-100 in PBS. Primary antibodies were: anti-PP1γ (1:300 dilution, sc-6108; Santa Cruz Biotechnology.); anti-CKII (1:100 dilution, 610444; BD Biosciences); anti-NEDD8 (1:100 dilution, ab50280; Abcam); anti-ZFHX4 (1:100 dilution, H00079776-A01; Novus Biologicals); and anti-α3 (1:300 dilution; Invitrogen). Primary antibodies were washed with PBS (3×), and the cells were incubated with Alexa 488 anti-rabbit IgG (2 μg/ml) and either Alexa 568 anti-mouse IgG1 (2 μg/ml) or Alexa 568 donkey anti-goat secondary antibodies (Molecular Probes) in 1% goat or donkey serum, 0.2% Triton X-100 in PBS for 1 h at room temperature. The secondary antibodies were washed with PBS (3×), and the cells were mounted using ProLong anti-fade mounting medium (Invitrogen). Single confocal sections were acquired with a laser scanning confocal microscope (60X, 1.4 NA, oil immersion) at ∼0.050 μm per pixel (Olympus FluoView or high resolution confocal built in-house). To improve spatial resolution, images were three-dimensional-blind deconvolved (AutoQuant Imaging Inc.). The degree of association between two proteins is measured via the protein proximity index (PPI) and correlation coefficients (CC), calculated using the recently published algorithms (15). The confocal data are representative images of four different mice preparations and 8–10 cells per preparation.
Proteasome Immunoprecipitation—
Purified 20 S proteasomes in Nonidet P-40 immunoprecipitation buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, protease inhibitor mixture from GE Healthcare) were incubated with rabbit anti-α3 antibody (2 μg antibody per 5 mg protein) and 10 μl of protein A/G-Sepharose resin (Santa Cruz Biotechnology) overnight at 4 °C. Samples were centrifuged at 1,000 × g for 2 min at 4 °C. The pellets were washed three times in Nonidet P-40 immunoprecipitation buffer and two times in PBS. Immunoprecipitated complexes were collected by centrifugation at 3,000 × g for 2 min at 4 °C. After removing the supernatant the proteasome-bound protein A/G beads were incubated in SDS-PAGE buffer for 5 min at 95 °C and then run on 12% SDS-PAGE gels. Gels were digested with trypsin and subsequently analyzed by LC-MS/MS.
20 S Proteasome Activity Assay—
The 20 S proteolytic activity assay was carried out in a total volume of 100 μl in 96-well opaque plates. The final composition of the 20 S assay buffer for the β5 proteasome activity was 25 mm HEPES, 0.5 mm EDTA, and 0.03% SDS (pH 7.5). Assays were initiated by addition of 10 μl of a 10× solution (250–5000 μm) of LLVY-AMC. The final composition of the 20 S assay buffer for the β1 and β2 proteasome activity were 25 mm HEPES, 0.5 mm EDTA, 0.05% Nonidet P-40, and 0.001% SDS (pH 7.5). Assays were initiated by addition of 10 μl of a 10× solution (250–5000 μm) of LLE-AMC (β1) or LSTR-AMC (β2). Released AMC was measured using a Fluoroskan Ascent fluorometer (Thermo Fisher Scientific) at excitation wavelength 390 nm and emission wavelength 460 nm. Each well contained 0.065 μg of purified 20 S proteasome. Fluorescence was measured at 15 min intervals for 2 h. All assays were linear in this range, and each sample was assayed in quadruplicate. A standard curve was generated using different concentrations of free AMC that was added to protein-free wells. Proteasome activity results were expressed as means ± S.E. For measuring proteasome activities in cytosolic fractions (1 h at 100,000 × g supernatants), controls were carried out using specific proteasome inhibitor epoxomicin or Z-Pro-Nle-Asp-H. Each well contained 25 μg of cytosolic fraction, and assays were carried out as described above for the purified proteasomes.
Activity assays were performed on purified heart or liver 20 S proteasomes incubated with 10 nm Calyculin A (PP1 inhibitor; Upstate) for 10 min or 100 nm DMAT (CKII inhibitor) for 20 min and measured for all three proteasome proteolytic activities 30 min after the addition of substrate. For the recombinant studies, 36 nm recombinant PP1 (NEB) or 15 units of CKII was added to purified proteasomes with proteolytic activity measured 30 min after the addition of β1, β2, or β5 substrates. Proteasomes incubated with recombinant PP1 or CKII inactivated by heating at 60 °C for 1 h were used as controls. Data are averages of at least three independent experiments. *, p < 0.05 for β5 cardiac activity with both recombinant PP1 compared with heat-inactivated recombinant PP1 and with inhibited PP1 compared with uninhibited activity. *, p < 0.05 for all three hepatic activities with recombinant CKII compared with the heat-inactivated recombinant CKII and inhibited CKII when compared with uninhibited activities.
Protein Identification by LC-MS/MS—
Protein concentrations were determined using the Bio-Rad Protein Assay Reagent, and protein samples were loaded at equal amounts and run with either two-dimensional electrophoresis (GE Healthcare) or BN-PAGE (as discussed above). Stained proteins and unstained regions within the vicinity (served as background control) were excised, reduced, and alkylated (16). After tryptic digestion, recovered peptide mixtures underwent LC-MS/MS analysis on an LTQ-MS instrument (Thermo Fisher Scientific, San Jose, CA) with a Surveyor pump system using a reversed phase column (75 μm, inner diameter, 10 cm; BioBasic C18, 5-μm particle size; New Objective, Woburn, MA). The flow rate was 5 μl/min for sample loading and 250 nl/min for separation. Mobile phase A was 0.1% formic acid, 2% acetonitrile in water, and mobile phase B was 0.1% formic acid, 20% water in acetonitrile. A shallow gradient was used for analyses: linear gradient from 5% B to 40% B over 70 min, then to 100% B over 20 min, and finally a constant 100% B for 9 min. The ion transfer tube of the linear ion trap was held at 200 °C; the normalized collision energy was 35% for MS2; and the spray voltage was set at 1.9 kV.
Survey full-scan MS spectra with 1 microscan (m/z 400–2000) were acquired, followed by five sequential scan events of MS2. Each MS2 acquisition was operated under the data-dependent acquisition mode to automatically select ions with the top five highest intensities from the survey scan, with a 3.0 m/z isolation width. During acquisition, dynamic exclusion was enabled with 2 repeat counts within 10.0 s and with exclusion duration of 40.0 s. For 18O labeling, the data were acquired similarly in a data-dependent mode with 1 full MS scan followed by MS2 collision-induced dissociation and zoom scans on the top five most intense precursor ions. The zoom scan mass width was set at ± 5 m/z.
MS/MS Spectra were searched against the international protein index mouse data base version 3.24 (which contains 52,326 proteins) with SEQUEST search engine (Bioworks 3.3). Fixed modifications were set for carbamidomethylation of cysteine. Differential modification was set for oxidation of methionine, as well as arginine and lysine mono- and di-methylated and allowing for one missed tryptic cleavage. Filter criteria were set to pass a significant threshold of cross-correlation versus charge state (2 for +1 ions, 2.2 for +2 ions, and 3.8 for +3 ions) and a probability threshold of 0.001. Two distinct peptides were required for all identified proteins. Relative quantitation of identified subunits was accomplished by taking the average peak areas of the two highest peaks for a given peptide. Peak intensities were normalized against the total peak intensities.
18O Labeling—
The post-digest 18O labeling procedure was performed similarly as described in the literature (17, 18). Briefly, the immobilized trypsin beads (Life Technology) were first washed by water and then added to the dried peptides of each tryptic digest (20% of the digest volume), and then again completely dried by SpeedVac. Either 18O-enriched water (95%; Sigma) or regular 16O water containing 20% acetonitrile was then each added to one of two samples and incubated overnight at 37 °C with gentle shaking. After labeling, the immobilized trypsin beads were removed using a microspin column and then further washed by acetonitrile. The 16O- or 18O-labeled samples were then combined and dried by SpeedVac. Samples were applied to the mass spectrometer as described previously. Results were the average of three independent experimental repetitions. The peaks were normalized to the median of all the ratios in the set of proteasome subunits.
RESULTS
We examined the composition, assembly, post-translational modifications, functional associating partners, and proteolytic function of the 20 S proteasomes from heart and liver (supplemental Fig. 1). The proteome biology and proteolytic functions of these two organs are summarized in supplemental Table 1. Proteome biology was studied using purified 20 S complexes (>98% purity); an approach combining multi-dimensional chromatography, and BN-PAGE was used (supplemental Fig. 2). The 20 S complexes visualized on BN-PAGE (Fig. 1A) were analyzed by MS/MS, identifying all 17 known proteasome components and the most abundant/easily detectable interacting proteins (supplemental Tables 2, 6, and 7). Our data suggests that associating proteins participated in these 20 S proteasomes in sub-stoichiometric amounts (consistent with other published reports) (19).
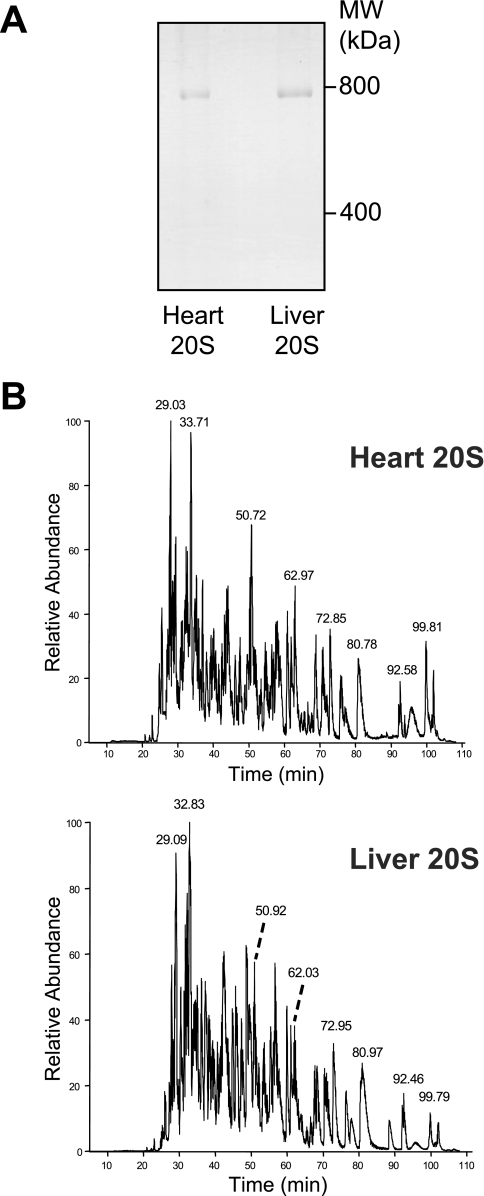
Fig. 1.
Analysis of purified murine cardiac and hepatic 20 S proteasomes. A, blue-native gel of purified murine cardiac and hepatic 20 S proteasomes. B, liquid chromatography of trypsin-digested heart and liver 20 S proteasome bands. The chromatograms for trypsin cleaved heart and liver 20 S proteasomes were similar, but distinct. Some of the major peaks in both samples are labeled.
Proteasome Composition and Assembly—
Results from BN-PAGE/LC-MS/MS show that the heart and liver 20 S proteasomes are distinct (Figs. 2–4).The chromatograms for the different trypsin-digested heart and liver 20 S samples differing in major components of the 20 S proteasomes (Fig. 1B). Three independent heart 20 S samples were remarkably similar with respect to the 20 S proteasome subunits identified and their sequence coverage (supplemental Table 2). The sequence coverage of these 17 known subunits ranged from 27.4% ± 2.9% (β1i) to 69.7% ± 2.9% (α1) for the heart, whereas those for the liver ranged from 40.9% ± 4.8% (β2i) to 64.7% ± 6.9% (β1).
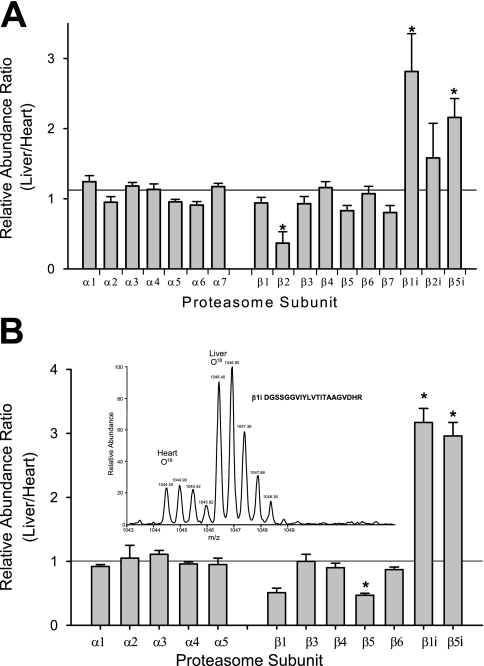
Fig. 2.
Quantification of heart and liver 20 S proteasome subunits. A, quantification using the average of the two highest peaks from the same peptides in liver and heart. n = 3; *, p < 0.05. B, quantification using 18O:16O labeling of proteasome subunits in the heart and liver. n = 3; *, p < 0.05. The peaks are normalized to the median of all the ratios in the set and are the result of three experimental repetitions. Inset shows a representative spectrum of 18O: 16O labeling of a β1i peptide.
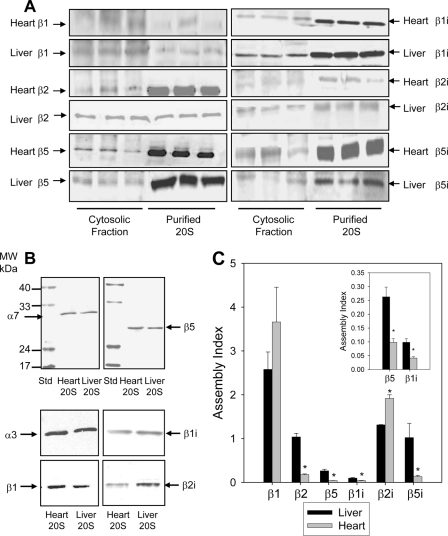
Fig. 3.
Comparison of free and assembled 20 S proteasome subunits in heart and liver by immunoblotting. A, comparison of constitutive and inducible proteasomes subunits in cytosolic fractions and purified 20 S from the heart and liver. Each lane contained 25 μg of cytosolic fraction or 1 μg of purified proteasome. B, comparison of proteasomes subunits in cytosolic fractions and purified 20 S from the heart and liver. Heart and liver 20 S proteasomes were run on SDS-PAGE, transferred to nitrocellulose, and probed with anti-proteasome antibodies. Each lane contained 1 μg of heart proteasome or 1 μg of liver proteasome. C, the assembly index for proteasome subunits in heart and liver cytosolic fractions. The assembly index is the ratio of the total amount of a proteasome subunit in the cytosolic fraction (free + partially assembled + assembled subunits) versus the amount of that proteasome subunit in the 20 S proteasome purified from the cytosolic fraction (only assembled subunits). The insert in Fig. 3C is an enlarged view of the assembly index for β5 and β1i.
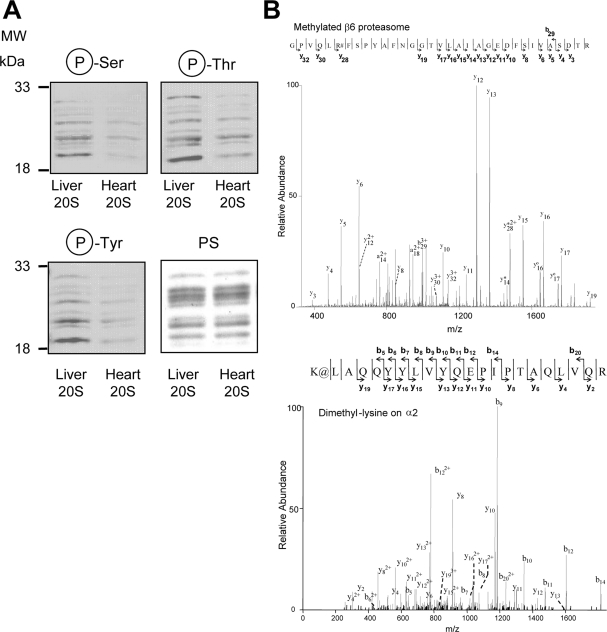
Fig. 4.
Comparison of post-translational modifications on purified 20 S proteasomes from heart and liver. A, comparison of the phosphoproteome of purified 20 S proteasomes from heart and liver. Each lane contained 2 μg of heart proteasome or 2 μg of liver proteasome. B, upper panel, mass spectra of a peptide from heart β6 proteasome subunit, which is mono-methylated on arginine; lower panel, a peptide from liver α2, which is dimethylated on lysine. # represents methylated arginine residue. The methylated peptide (β6) had an Xcorr of 4.114 and a precursor ion m/z of 1157.30 (3+ charge). @ represents dimethylated lysine residue. The dimethylated peptide (α2) had an Xcorr of 5.266 and a precursor ion m/z of 893.23 (3+ charge). Protein loading was controlled by Ponceau S stain (PS).
Three different approaches were used to quantitatively determine the assembly of three β subunits (β1, β2, β5) and their three inducible counter parts (β1i, β2i, β5i) in the heart and liver 20 S proteasomes. First, quantification of the heart and liver 20 S proteasome subunits was carried out using the average peak area of the two highest peaks of a given peptide detected in both the heart and liver 20 S proteasomes. Label-free LC-MS/MS quantification of the 20 S proteasomes from heart and liver suggested significantly higher amounts of inducible subunits (β1i, β5i) in liver when compared with heart (p < 0.05, n = 3) (Fig. 2A). Second, using 16O:18O post-digestion labeling of the heart and liver proteasomes (Fig. 2B and supplemental Table 3), we also observed significantly lower inducible β subunits in the heart. Using this 16O: 18O labeling the concentration of liver/heart for β1i and β5i were 3.17 ± 0.22 and 2.96 ± 0.21, respectively. Finally, a third approach was used to investigate the potentially higher levels of inducible β catalytic subunits in the liver using immunoblotting. Odyssey infrared imaging of the purified 20 S proteasomes shown in Fig. 3B allows for the quantification of the level of a given subunit.
By comparing the level of a given β catalytic subunit in the cytosol to the level in purified samples, we obtain a ratio that is directly comparable between tissues. The ratio of the total proteasome subunits (obtained from cytosolic fractions) versus the proteasome subunits fully integrated into mature 20 S complexes (obtained from purified 20 S proteasomes) is referred to as the “assembly index” (Fig. 3, A and C and supplemental Table 5). These ratios, or the assembly index, compares the capacity of the heart and liver to incorporate subunits into mature proteasomes. The assembly index values for β2, β5, β1i, and β5i were greater in liver than in heart, with β2i as the exception. This provides further evidence that the liver 20 S proteasomes maintain a higher level of inducible β subunits in the liver (Fig. 3 and supplemental Table 5). Other subunits were also compared (α3, α5, and α7), and although the liver contains a higher amount of α subunits, no significant difference between the levels of α3, α5, and α7 subunits in the heart relative to the liver cytosolic fractions (supplemental Fig. 8) were observed. This suggests that there is no substantial variability in these proteasome subunit expression levels between the heart and liver.
Post-translational Modifications—
Immunoblotting showed that the liver 20 S proteasomes contained higher relative amounts of phosphorylated Ser, Thr, and Tyr residues than the cardiac 20 S proteasomes (Fig. 4A), which is consistent with our previous findings (11). We have recently shown that the heart proteasome is phosphorylated on at least six residues and that phosphorylation is a key regulator of the proteasome (13, 10). Using immunoblotting, the presence of several post-translational modifications on proteasomes were investigated: ubiquitination, sumoylation, tyrosine nitrosation, gluthathionolation, neddylation, and methylation. Although the proteasome fraction was found to be neddylated, no proteasome subunits were neddylated based upon the molecular mass of the neddylated protein (Fig. 5). Using mass spectrometry and immunoblotting we provide the first evidence of proteasome methylation (Fig. 4B and supplemental Fig. 5). The β6 proteasome subunit was found to be monomethylated on arginine, whereas α2 was found to be dimethylated on lysine (Fig. 4B). Immunoblotting of heart and liver proteasomes on native and two-dimensional gels using methyl-specific antibodies suggest that some proteasome subunits are methylated (supplemental Fig. 5). The purified 20 S proteasome was not found to be ubiquitinated, sumoylated, lysine-acetylated, or glutathiolated (supplemental Fig. 6).
Fig. 5.
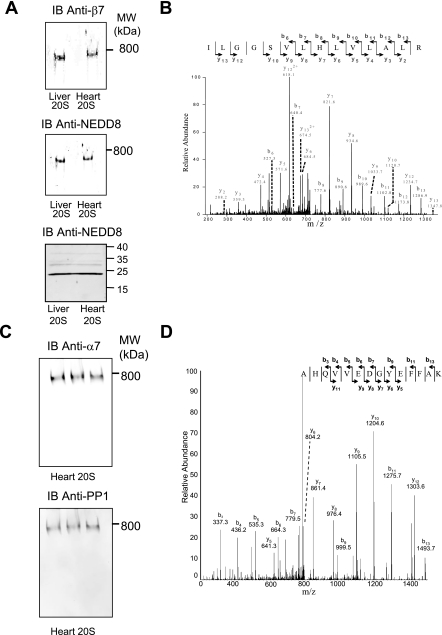
Validation of associating partners of cardiac and hepatic 20 S proteasomes. A, cardiac and hepatic 20 S proteasome was run on a native gel, transferred to nitrocellulose, and probed with polyclonal anti-NEDD8 and monoclonal anti-β7. B, identification of a peptide from NEDD8 present in the BN-PAGE 20 S band using LC-MS/MS analysis. C, immunoblot of native gel to verify interaction of PP1 with the heart 20 S proteasomes. D, identification of a peptide from PP1 present in the BN-PAGE 20 S band using LC-MS/MS analysis.
Functional Associating Partners—
Previous studies on proteasome associating partners have proven successful (20, 21). In our studies, the native 20 S complexes displayed by the BN-PAGE were analyzed by MS/MS for associating partner identification. Nine different associating partners were found to stably interact with the native 20 S proteasome complexes in both the heart and the liver including elongation factor 2, 90 kDa heat shock protein, stress-70 protein mitochondrial precursor and calpain 2 catalytic subunit (spectra shown in supplemental Fig. 4). Fig. 5 shows the MS/MS for one of the peptides used to identify NEDD8 as an associating partner for the 20 S proteasome. Several pilot experiments were carried out to assure appropriate separation of multi-protein complexes on the BN-PAGE. Comparison of proteins identified with the 20 S complexes to those of the non-20 S complexes (those running below or above the 20 S proteasome band) enabled the elimination of false positive proteins such as glial fibrillary acidic protein.
Immunoblotting also demonstrated that NEDD8 was part of the 20 S proteasome complexes (Fig. 5). Several other well characterized proteasome interacting partners, protein kinase A (PKA) and protein phosphatase 2A (PP2A) (10), as well as a known interacting partner, CKII, and a previously unknown interacting partner, PP1, were all observed by mass spectrometry and verified by immunoblotting (data not shown). Fig. 5 shows a spectrum from PP1 obtained from the native cardiac 20 S proteasomes, which was confirmed by immunoblotting. Similar results were also obtained for liver 20 S proteasomes (data not shown). Most of the interacting partners were found to be present in both heart and liver 20 S samples. The 11 S regulatory subunits were also examined (supplemental Fig. 9). Although no significant amount of 11 Sγ was found in either cytosolic proteasomes, the 11 Sα and 11 Sβ were significantly more abundant in the liver than in heart cytosol.
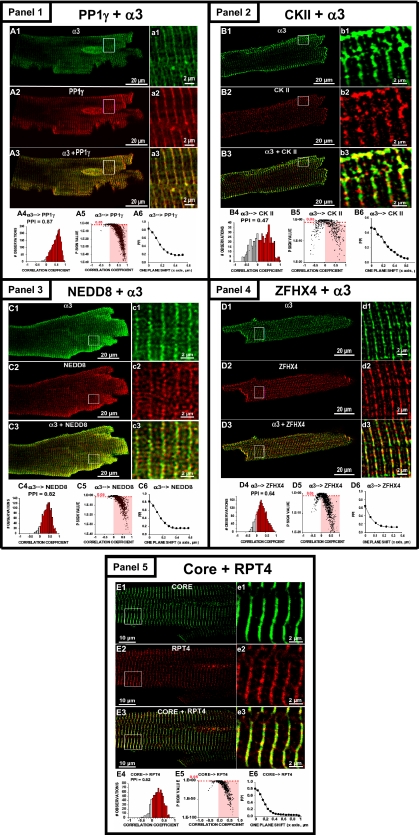
Additional confidence for the verification of associating partners was demonstrated via high resolution confocal microscopy and co-immunoprecipitation. To gain insight into the cellular distribution and binding of associating partners to the proteasomes in mammalian cells, we immunostained isolated mammalian cells and imaged with confocal microscopy. PP1, CKII, NEDD8, and ZFHX4 were all confirmed to colocalize with the proteasome complexes, providing additional confidence that the associating partners were identified through BN-gel (Fig. 6). The degree of association between two proteins was measured using the PPI and CC. The PPI and CC are independent measures for validating the colocalization of PP1, CKII, NEDD8, and ZFHX4 with the proteasome and are consistent with values obtained for the colocalization of the 19 S complex (Rpt4) with the core 20 S proteasome (Fig. 6, panel 5). Associating proteins were also verified by co-immunoprecipitating the core 20 S complex from purified 20 S proteasome preparations and subsequent MSn identification of non-proteasome proteins. Several of the associating proteins, including PP1, CKII, PKA, and PP2A were still detected to associate with 20 S proteasome complexes after co-immunoprecipitation (supplemental Table 8). Identification of associating proteins through a combination of BN-gel, immunohistochemistry, and co-immunoprecipitation provides considerable confidence that the interactions are biologically relevant.
Fig. 6.
Confocal validation of the associating partners with proteasome complexes in mammalian cells. Panel 1, representative single confocal sections of cardiomyocytes immunostained with anti-20 S proteasome α3 (A1, green), anti-PP1γ (A2, red), and the overlay of α3 and PP1γ (A3). a1–a3 are the regions in the squares at higher display magnification. A4, CC histogram. PPI of 0.87 for α3 with proximity to PP1γ, indicating a high degree of association. A5, P significance test versus CC for α3 with proximity to PP1γ; the highlighted area has <0.05. A6, PPI as a function of pixel shift. Panel 2, immunostained cells with anti-20 S proteasome α3 (B1, green), anti-CKII (B2, red), and overlay (B3). b1–b3 are regions at higher magnification. B4, CC histogram. PPI of 0.47 for α3 proximity to CKII. B5, CC for α3 with proximity to CKII; the highlighted area has p < 0.05. B6, PPI as a function of pixel shift. Panel 3, immunostained cells with anti-20 S proteasome α3 (C1, green), anti-NEDD8 (C2, red), and overlay (C3). c1–c3 are regions at higher magnification. C4, CC histogram. PPI of 0.82 for α3 proximity to NEDD8. C5, CC for α3 with proximity to NEDD8; the highlighted area has p < 0.05. C6, PPI as a function of pixel shift. Panel 4, immunostained cells with anti-20 S proteasome α3 (D1, green), anti-ZFHX4 (D2, red), and overlay (D3). d1–d3 are regions at higher magnification. D4, CC histogram. PPI of 0.64 for α3 with proximity to ZFHX4. D5, CC for α3 proximity to CKII; the highlighted area has p < 0.05. D6, PPI as a function of pixel shift. Immunostained cells with anti-core 20 S proteasome (E1, green), anti-Rpt4 19 S proteasome (E2, red), and overlay of core and Rpt4 (E3). e1–e3 are regions in the squares at higher magnification. E4, CC histogram. PPI of 0.82 for core proximity to Rpt4. E5, Psign test versus CC for core proximity to Rpt4; the highlighted area has p < 0.05. E6, PPI as a function of pixel shift.
Proteasome Activities—
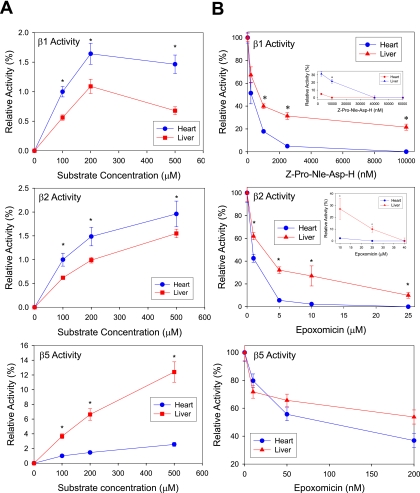
The three proteolytic activities (β1 caspase-like, β2 tryptic-like, β5 chymotryptic-like) of the cytosolic 20 S proteasomes were examined (Fig. 7A). Distinct patterns in proteasome function of these two organs were observed: The chymotrypsin-like proteolytic activity of the hepatic cytosolic 20 S proteasomes was significantly greater than that from cardiac cytosolic 20 S proteasomes, whereas the caspase and trypsin-like proteolytic activities of the cardiac cytosolic 20 S proteasomes was greater than that of hepatic cytosolic 20 S proteasomes.
Fig. 7.
Proteolytic activities of the purified 20 S proteasomes from the heart and liver. A, comparison of the proteolytic activities of the 20 S proteasomes in cytosolic fractions from the heart and liver. *, p < 0.05; n = 4. B, comparison of the inhibition of 20 S proteasome activities in cytosolic fractions from the heart and liver. *, p < 0.05; n = 4. Heart and liver cytosol fractions (25 μg each) were assayed for β1, β2, and β5 activities in the presence of 100–500 mm of fluorescent substrate. *, p < 0.05; n = 5. C, comparison of the proteolytic activities of the purified 20 S proteasomes from the heart and liver. *, p < 0.05; n = 4. D, comparison of the inhibition of purified 20 S proteasome activities from the heart and liver.
To evaluate tissue heterogeneity and sensitivity to proteasome inhibition we examined the effect of two proteasome inhibitors on the proteolytic function of heart and liver. Our data showed that the heart had a higher susceptibility to epoxomicin than liver (Fig. 7B). Investigation of the purified heart and liver 20 S proteasomes showed that the caspase-like and trypsin-like proteolytic activities were greater in the liver (Fig. 7C). The effect of proteasome inhibitors on the purified heart and liver 20 S complexes was also investigated (Fig. 7D). The heart 20 S proteasomes were highly sensitive to epoxomicin inhibition when compared with liver as shown in their respective β5 chymotrypsin-like activity.
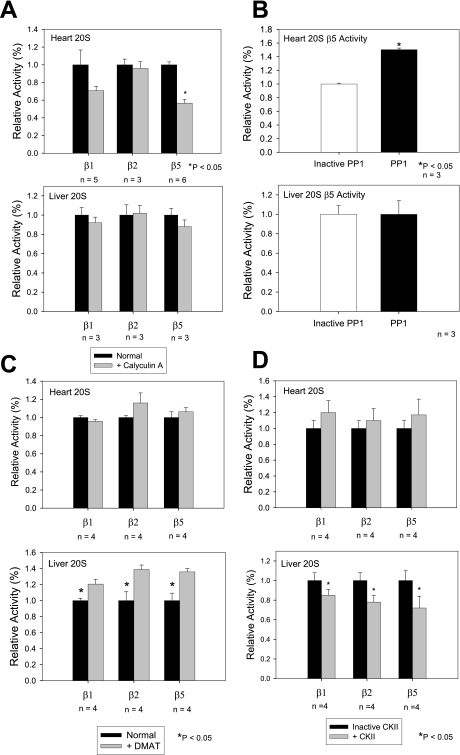
PP1, which was associated with heart and liver 20 S complexes, showed distinct functional roles in the heart and liver (Fig. 8). Addition of PP1 to cardiac 20 S proteasomes significantly enhanced the β5 chymotrypsin-like activity (50% ± 2% increase when compared with heat-inactivated PP1); whereas PP1 had no detectable effect on the β5 chymotrypsin-like activity of the liver 20 S proteasomes (Fig. 8B). Inhibition of the endogenous CKII bound to 20 S proteasomes resulted in an increase in all three proteolytic activities of the liver proteasome without affecting the cardiac proteasome (Fig. 8C). Addition of CKII to liver 20 S proteasomes significantly decreased all three proteolytic activities of the liver proteasome without affecting the cardiac proteasome (Fig. 8D).
Fig. 8.
Characterization of the effects of calyculin A, PP1, DMAT (CKII inhibitor), or CKII on the proteolytic activities of the purified proteasomes. A, proteolytic activities of purified 20 S proteasomes 50 min after the addition of 10 nm PP1 inhibitor calyculin A. Data is average over 3–6 experiments. B, β5 proteolytic activity 50 min after 36 nm recombinant PP1 added to purified 20 S proteasomes. Other proteolytic activities showed no detectable effect. Data is average of three experiments. *, p < 0.05. C, proteolytic activities of purified 20 S proteasomes 50 min after addition of 100 nm CKII inhibitor DMAT. D, β5 proteolytic activity 60 min after 15 units of recombinant CKII were added to purified 20 S proteasomes. *, p < 0.05; n = 3–6 for all experiments.
DISCUSSION
This investigation identified contrasting alterations of proteasome inhibition in the liver versus the heart. The sensitivities of the cytosolic cardiac 20 S proteasomes to the two proteasome inhibitors tested (epoxomicin and Z-Pro-Nle-Asp-H) were dramatically higher than in the liver. These functional consequences are concomitant with the differential proteome biology of 20 S proteasomes observed in these two organs. Despite identical genetics, 20 S complexes of these two organs differed in their molecular composition, complex assembly, post-translational modifications, functional associating partners, regulatory complexes, and proteolytic function. To our knowledge, this is the first investigation that delineates the tissue-specific heterogeneity of 20 S proteasomes in the same animal strain.
Molecular Composition and Assembly—
Free proteasome subunits, partially assembled proteasomes and fully assembled proteasomes all exist simultaneously within the cell. The assembly and molecular organization of the proteasome is complex, as two sets of 7α and 7β subunits are required for 20 S proteasome assembly. The three β subunits (β1, β2, β5) can be replaced with their inducible counterparts (β1i, β2i, β5i), all of which possess catalytic activity. We have previously shown the parallel integration of all six β subunits in the 20 S proteasomes from normal adult myocardium (13, 10). In this report, using three independent experimental methods (label free quantification, 16O:18O labeling and immunoblotting), we show that the six β subunits of both cardiac and hepatic 20 S proteasomes are assembled in distinct ratios and patterns. Regulatory mechanisms for proteasome assembly in the heart and liver remain to be determined. A recent paper by Sharon et al. (22) showed that the assembly of β subunits may involve propeptide processing. The degradation of proteasome subunits is also unknown and may involve the release of previously assembled proteasome subunits into the cytosol for degradation.
Post-translational Modifications—
Multiple forms of post-translational modifications of proteasomes have been reported. In this investigation and previous studies from our group (10, 11, 9) we have identified phosphorylation, N-terminal acetylation, myristoylation, and arginine and lysine methylation of proteasome subunits. These results were validated with a combined approach using mass spectrometry and immunoblotting with specific antibodies. Of interest is the detection of arginine and lysine methylation in both heart and liver 20 S subunits. This form of post-translational modification has been implicated in other cellular processes, including protein trafficking, signal transduction, and transcriptional regulation (23). Arginine methylation is seen as a common post-translational modification and is important for protein-protein interactions. More methylated proteins are likely to be discovered in the near future as predicated by the high percentage of genes in the mammalian genome (over 1%) that encode methyltransferases (24). This is the first report of methylated proteasome subunits. The specific role of methylation on proteasome subunits is unknown but could be important for the interaction with their associating partners.
Functional Associating Partners—
The proteasomes fulfill a wide range of functions in the eukaryotic cell and it is anticipated that 20 S proteasomes interact with different proteins that could modulate its activity. Any two protein associations are governed by the level of expression and degradation, the ratio of free versus assembled/associated, and the subpopulation of protein with independent function(s) from its associated/assembled form. To date, many proteasome interacting proteins are currently identified (23). These 20 S complex associated proteins may be substrates, accessory proteins, or enzymes associated with 20 S proteasome function. In this study we used blue-native gels to characterize 20 S proteasome associating partners. BN-PAGE allows for high-resolution separation of multi-protein complexes under native conditions. The final electrophoretic mobility of a multiprotein complex is determined by the amount of Coomassie dye bound and the size and shape of the complex (25).
In this study, we report two proteasome associating proteins (NEDD8 and elongation factor 2) that were found in all six 20 S proteasome preparations (three cardiac and three hepatic preparations). NEDD8, also called neddylin, is a ubiquitin-like protein (80% homology to ubiquitin) (26), that is activated by the E1 ubiquitin-activating enzyme UBA2 and then linked to an E2-like enzyme, UBC12. Like ubiquitin, NEDD8 is attached covalently to targeted proteins (neddylation); however, the biological function of NEDD8 is unclear. NEDD8 play important roles in development and differentiation, possibly regulating the E3 ubiquitin ligase complex SCF (Skp1/Cul1/Cdc53 F box) (27), and possibly playing a role in proteolysis via the ubiquitin-proteasome system (28, 29). Elongation factor 2 is a cytosolic protein that promotes the GTP-dependent translocation of the nascent protein chain from the A-site to the P-site of the ribosome. Its interaction with the 20 S proteasome agrees well with recent reports that Elongation factor 1 interacts with proteasomes (30). Although the interactions between elongation factor 2 and 20 S proteasomes have not been previously detected, it is likely that these translational elongation factors are involved in the degradation of co-translationally damaged proteins, linking protein synthesis and degradation pathways (31).
Other proteins that were found to associate with the proteasome in more than one 20 S proteasome sample include: protein kinase A (10), protein phosphatase 2A (10), casein kinase II, protein phosphatase 1, calpain 2, stress-70 protein mitochondrial precursor, 90 kDa heat shock protein, arginine N-methyl transferase 1, and annexin A2 (supplemental Fig. 4). The presence of casein kinase II, protein phosphatase 1, heat shock protein 90, protein kinase A, protein phosphatase 2A, elongation factor 2, and NEDD8 in both the heart and liver (Fig. 5) contrasts with the existence of zinc finger homeodomain 4 solely in the heart (supplemental Fig. 7). The inevitable loss of the more weakly bound or transient proteins during purification highlights the dynamism of the proteasome complex as well as the difficulty in developing a comprehensive map of associating partners. A growing list of 20 S proteasome associating partners, both shared and unique to particular organs, enhances the potential of functional diversity of the proteasome complexes.
Proteolytic Activity—
The differences in the proteolytic activities between the heart and liver may be attributed by an organ-specific proteasome subunit composition and/or a difference in associating partners. It is also possible that because of complex heterogeneity, proteasome activators or inhibitors may differently affect proteasome activities of the heart and liver. In our study, the unique proteolytic substrate pattern exhibited by the cardiac cytosolic 20 S (higher β1 and β2 and lower β5 activities) suggests complex multifaceted mechanisms in regulating the proteasome activity in a tissue-specific manner. PP1 was associated with both the murine cardiac and hepatic 20 S proteasomes. Exogenous PP1 affected the 20 S proteasome in a tissue-specific role as it increased β5 proteolytic activity in the heart, but had no significant effect in liver. The PP1 inhibitor, calyculin A, decreased β5 activity without a demonstrable effect on β1 and β2 proteolytic activities. Inhibition of the endogenous CKII within 20 S proteasome complexes resulted in an organ-specific effect on the liver proteolytic activities. CKII has been previously shown to phosphorylate α3 and α7 in vitro (32). Addition of CKII to liver 20 S proteasome significantly decreased all three proteolytic activities, whereas CKII did not affect the cardiac 20 S proteasome.
Conclusion—
This investigation shows that despite identical genetic input, the heart and liver manifested contrasting proteome biology in their 20 S proteasome complexes. The differences in the molecular assembly of the catalytically active β subunits as well as distinct associating partners contributed to the differential proteolytic consequences that these two organs possess. These data illustrate that structural and functional features of 20 S proteasomes are heterogeneous between tissue types. Furthermore, the cardiac proteasomes displayed much higher sensitivity to the drug epoxomicin than that of the liver. This may explain why the proteasome inhibitor Velcade showed more toxicity to cardiac cells than to normal colon cells (33). The ubiquitous presence of the 20 S proteasomes in all tissue types and the heterogeneity identified in this study recommends caution to the application of systematic administration of proteasome inhibitors in future therapeutic regiments.
Supplementary Material
Footnotes
Published, MCP Paper in Press, October 17, 2008, DOI 10.1074/mcp.M800058-MCP200
The abbreviations used are: BN-PAGE, blue-native polyacrylamide gel electrophoresis; DTT, dithiothreitol; PBS, phosphate-buffered saline; PPI, protein proximity index; CC, correlation coefficients; PKA, protein kinase A; PP2A, protein phosphatase 2A; CKII, casein kinase II; PP1, protein phosphatase 1; LC-MS/MS, liquid chromatography tandem mass spectrometry; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; AMC, 7-amino-4-methyl coumarin.
This work was supported, in whole or in part, by National Institutes of Health Grant HL R01–63901, HL R01–65431, and HL P01-80111 (to P. P.), HL BRG 088640 (to E. S.), AHA 0160144B (to A. G.), and AHA 0715004Y (to G. Y.). This work was also supported by an endowment from Theodore C. Laubisch (to P. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Merchionne, F., Perosa, F., and Dammacco, F. ( 2007) New therapies in multiple myeloma. Clin. Exp. Med. 7, 83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voortman, J., and Giaccone, G. ( 2006) Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: a case report. BMC Cancer 6, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enrico, O., Gabriele, B., Nadia, C., Sara, G., Daniele, V., Giulia, C., Antonio, S., and Mario, P. ( 2007) Unexpected cardiotoxicity in haematological bortezomib treated patients. Br. J. Haematol. 138, 396–397 [DOI] [PubMed] [Google Scholar]
- 4.Hacihanefioqlu, A., Tarkun, P., and Gonulle, E. ( 2008) Acute severe cardiac failure in myeloma patient due to proteasome inhibitor bortezmib. Int. J. Hematol. 88, 219–222 [DOI] [PubMed] [Google Scholar]
- 5.Asher, G., Bercovich, Z., Tsvetkov, P., Shaul, Y., and Kahana, C. ( 2005) 20 S proteasomal degradation of ornithine decarboxylase is regulated by NQO1. Mol. Cell 17, 645–655 [DOI] [PubMed] [Google Scholar]
- 6.Asher, G., Tsvetkov, P., Kahana, C., and Shaul, Y. ( 2005) A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 19, 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glickman, M. H., and Ciechanover, A. ( 2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 8.Sijts, A., Zaiss, D., and Kloetzel, P. M. ( 2001) The role of the ubiquitin-proteasome pathway in MHC class I antigen processing: implications for vaccine design. Curr. Mol. Med. 1, 665–676 [DOI] [PubMed] [Google Scholar]
- 9.Young, G. W., Wang, Y., and Ping, P. ( 2008) Understanding proteasome assembly and regulation: importance to cardiovascular medicine. Trends Cadiovasc. Med. 3, 93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zong, C., Gomes, A. V., Drews, O., Li, X., Young, G. W., Berhane, B., Qiao, X., French, S. W., Bardag-Gorce, F., and Ping, P. ( 2006) Regulation of murine cardiac 20 S proteasomes: role of associating partners. Circ. Res. 99, 372–380 [DOI] [PubMed] [Google Scholar]
- 11.Drews, O., Wildgruber, R., Zong, C., Sukop, U., Nissum, M., Gomes, A. V., and Ping, P. ( 2007) Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol. Cell. Proteomics 6, 2021–2031 [DOI] [PubMed] [Google Scholar]
- 12.Brookes, P. S., Pinner, A., Ramachandran, A., Coward, L., Barnes, S., Kim, H., and Darley-Usmar, V. M. ( 2002) High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signaling complexes. Proteomics 2, 969–977 [DOI] [PubMed] [Google Scholar]
- 13.Gomes, A. V., Zong, C., Edmondson, R. D., Li, X., Stefani, E., Zhang, J., Jones, R. C., Thyparambil, S., Wang, G. W., Qiao, X., Bardag-Gorce, F., and Ping, P. ( 2006) Mapping the murine cardiac 26 S proteasome complexes. Circ. Res. 99, 362–371 [DOI] [PubMed] [Google Scholar]
- 14.Eghbali, M., Deva, R., Alioua, A., Minosyan, T. Y., Ruan, H., Wang, Y., Toro, L., and Stefani, E. ( 2005) Molecular and functional signature of heart hypertrophy during pregnancy. Circ. Res. 96, 1208–1216 [DOI] [PubMed] [Google Scholar]
- 15.Ropero, A. B., Eghbali, M., Minosyan, T, Y., Tang, G., Toro, L., and Stefani, E. ( 2006) Heart estrogen receptor alpha: distinct membrane and nuclear distribution patterns and regulation by estrogen. J. Mol. Cell. Cardiol. 41, 496–510 [DOI] [PubMed] [Google Scholar]
- 16.Bruneel, A., Labas, V., Mailloux, A., Sharma, S., Royer, N., Vinh, J., Pernet, P., Vaubourdolle, M., and Baudin, B. ( 2005) Proteomics of human umbilical vein endothelial cells applied to etoposide-induced apoptosis. Proteomics 15, 3876–3884 [DOI] [PubMed] [Google Scholar]
- 17.An, Y., Fu, Z., Gutierrez, P., and Fenselau, C. ( 2005) Solution isoelectric focusing for peptide analysis: comparative investigation of an insoluble nuclear protein fraction. J. Proteome Res. 4, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18.Qian, W. J., Monroe, M. E., Liu, T., Jacobs, J. M., Anderson, G. A., Shen, Y., Moore, R. J., Anderson, D. J., Zhang, R., Calvano, S. E., Lowry, S. F., Xiao, W., Moldawer, L. L., Davis, R. W., Tompkins, R. G., Camp, D. G., 2nd and Smith, R. D. ( 2005) Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol. Cell. Proteomics 4, 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coux, O., Tanaka, K., and Goldberg, A. L. ( 1996) Structure and functions of the 20 S and 26 S proteasomes. Annu. Rev. Biochem. 65, 801–847 [DOI] [PubMed] [Google Scholar]
- 20.Leggett, D. S., Hanna, J., Borodovsky, A., Crosas, B., Schmidt, M., Baker, R. T., Walz, T., Ploegh, H., and Finley, D. ( 2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 21.Wang, X., and Huang, L. ( 2008) Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol. Cell. Proteomics 7, 46–57 [DOI] [PubMed] [Google Scholar]
- 22.Sharon, M., Witt, S., Glasmacher, E., Baumeister, W., and Robinson, C. V. ( 2007) Mass spectrometry reveals the missing links in the assembly pathway of the bacterial 20 S proteasome. J. Biol. Chem. 282, 18448–18457 [DOI] [PubMed] [Google Scholar]
- 23.Bedford, M. T., and Richard, S. ( 2005) Arginine methylation: an emerging regulator of protein function. Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 24.Katz, J. E., Dlakic, M., and Clarke, S. ( 2003) Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteomics 2, 525–540 [DOI] [PubMed] [Google Scholar]
- 25.Schagger, H., Cramer, W. A., and von Jagow, G. ( 1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 26.Kamitani, T., Kito, K., Nguyen, H. P., and Yeh, E. T. ( 1997) Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 272, 28557–28562 [DOI] [PubMed] [Google Scholar]
- 27.Kawakami, T., Chiba, T., Suzuki, T., Iwai, K., Yamanaka, K., Minato, N., Suzuki, H., Shimbara, N., Hidaka, Y., Osaka, F., Omata, M., and Tanaka, K. ( 2001) NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamitani, T., Kito, K., Fukuda-Kamitani, T., and Yeh, E. T. ( 2001) Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J. Biol. Chem. 276, 46655–46660 [DOI] [PubMed] [Google Scholar]
- 29.Tateishi, K., Omata, M., Tanaka, K., and Chiba, T. ( 2001) The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokumoto, T., Kondo, A., Miwa, J., Horiguchi, R., Tokumoto, M., Nagahama, Y., Okida, N., and Ishikawa, K. ( 2003) Regulated interaction between polypeptide chain elongation factor-1 complex with the 26 S proteasome during Xenopus oocyte maturation. BMC Biochem. 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuang, S. M., Chen, L., Lambertson, D., Anand, M., Kinzy, T. G., and Madura, K. ( 2005) Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol. Cell. Biol. 25, 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castano, J. G., Mahillo, E., Arizti, P., and Arribas, J. ( 1996) Phosphorylation of C8 and C9 subunits of the multicatalytic proteinase by casein kinase II and identification of the C8 phosphorylation sites by direct mutagenesis. Biochemistry 35, 3782–3789 [DOI] [PubMed] [Google Scholar]
- 33.Papandreou, C. N., Daliani, D. D., Nix, D., Yang, H., Madden, T., Wang, X., Pien, C. S., Millikan, R. E., Tu, S. M., Pagliaro, L., Kim, J., Adams, J., Elliott, P., Esseltine, D., Petrusich, A., Dieringer, P., Perez, C., and Logothetis, C. J. ( 2004) Phase I trial of the proteasome inhibitor bortezomib in patients with advanced solid tumors with observations in androgen-independent prostate cancer. J. Clin. Oncol. 22, 2108–2121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.