Abstract
Phosphoinositide phosphates, PtdInsP, are important components of the cell lipid pool that can function as messengers in diverse cellular processes. Lack of information on downstream targets, however, has impeded our understanding of the potential of lipid-signaling to influence gene activity. Our goals here were to identify genes that altered expression in the presence of two isomeric monophosphate lipid messengers (Phosphoinositide 5-Phosphate, PtdIns(5)P, and Phosphoinositide 4-Phosphate, PtdIns(4)P) and to establish whether the two lipids influence distinct or overlapping gene-sets. Our results indicated that PtdIns(5)P and PtdIns(4)P affected genes within shared gene-families but that each messenger influenced the expression of different members within the same family. These results suggested that PtdIns(5)P and PtdIns(4)P participate in separate pathways that, ultimately, may control gene expression. The pathways may have points of convergence but may also counteract each other's effects. A significant fraction (∼40%) of the PtdIns(5)P-stimulated genes belong to various families of wall-modifying genes. Wall-modifying activities are recognized as factors affecting cell extension and plant growth. Elevated PtdIns(5)P concentration influenced stem growth and the effects were different from those triggered by PtdIns(4)P. The data allow insights into plants' response to two related PtdInsP at whole-plant/genome-wide levels and demonstrate that PtdIns(5)P-and PtdIns(4)P-involving mechanisms are distinct, selective and specific.
Key Words: Arabidopsis, microarrays, phosphoinositides, signaling pathways, genome response
Introduction
Lipid signaling mechanisms mediating growth, development, and responses to biotic and abiotic stresses are becoming an increasingly attractive topic of current plant research.1 Phasphatidyl-inositide phosphates, PtdInsPs, for example, may control a variety of processes required for normal cellular functions including cytoskeletal rearrangements, membrane trafficking and signal transduction.1–3 Yeast, animal, and plant cells respond to hyperosmotic stress by rapidly increasing the synthesis of phosphatidyl inositol-bisphosphates PtdIns(3,5)P2 and PtdIns(4,5)P2.4–6 In addition to bisphosphates, monophosphorylated isomers, PtdIns(3)P, PtdIns(4)P, and PtdIns(5P), are also implicated in cell signaling processes.7 In mammalian cells, (PtdIns(4)P) is important for membrane biogenesis and vesicle trafficking from the ER to the Golgi and plasma membranes;8 PtdIns(5)P is a minor but distinct component of the lipid pool, a main intermediate in the cell osmoprotective response pathway,9 and an important factor in the etiology of myotubular myopathy.10
Plants and animals share common aspects of lipid signaling, although plant-unique features have been recognized (reviewed in ref. 1). In plant cells, PtdIns(4)P was found widely distributed in various subcellular compartments suggesting that the lipid might be involved in distinct physiological roles.11 PtdIns(3)P and PtdIns(4)P regulate stomatal aperture and ABA-induced Ca2+ signaling.12 Unknown to exist in plants until recently, PtdIns(5)P was detected as a compound increasing its levels in response to hyperosmotic stress.5 The data suggested that lipid molecules, PtdInsP in particular, may act as second messengers in a wide scope of processes critical for cell survival, environmental adaptation and growth. The phosphoinositides can penetrate both hydrophilic and hydrophobic environments, making them messengers that may travel between, and within, cells. Existence of diverse phosphorylated isomers creates selective means for communication and for coordinating cell growth (reviewed in ref. 11).
Various mechanisms were proposed to translate the signal carried by the lipid messenger into forces that would promote cell response.13 However, lack of knowledge of downstream targets of lipid-involving mechanisms has impeded understanding of their potential to control gene functions. Crosstalk and overlaps of different signaling processes1,14 further complicate the picture, underscoring the necessity to outline distinct pathways and to define ultimate targets. Comparative analyses of transcription profiling, reflecting changes induced by various stresses, could provide a strategy to locate points of convergence-divergence within networks.14
It is becoming increasingly evident that lipids may be involved in broader functions than signalling linked exclusively with the plasma membrane.15 For example, PtdIns(4,5)P2 was found in heterochromatin, in a complex with BAF, and involved in pre-mRNA processing (reviewed in ref. 7). PtdIns(5)P regulates the tumor suppressor ING216 and inositol polyphosphates can bind chromatin modifying complexes.17,18 The ARABIDOPSIS HOMOLOG OF TRITHORAX, ATX1, a plant epigenetic regulator controlling growth, formation, placement, and identity of flower organs19 bound specifically PtdIns(5)P suggesting a novel signaling pathway in Arabidopsis.20 These newly emerging ideas, involving lipid signaling in gene regulatory mechanisms, make the conceptual framework for the study reported here. As part of our ongoing effort to determine factors that regulate Arabidopsis genome activity, we identified potential gene-targets of a putative PtdIns(5)P-ATX1 signaling mechanism.20 Here, in a genome-wide expression profiling, we report genes that alter their transcription in response to elevated PtdIns(5)P-levels. The specificity of the response is illustrated by the genome-wide analysis and identification of a distinct genes changing expression in response to a different lipid monophosphate, PtdIns(4)P. Cluster analyses revealed that the Arabidopsis genome responded selectively to each of the two monophosphate isomers and that PtdIns(5)P and PtdIns(4)P may use different pathways to influence their targets. The physiological relevance of the gene targets established by the microarrays is experimentally demonstrated for a group of functionally related (wall-modifying) genes that were upregulated by elevated PtdIns(5)P, but not PtdIns(4)P. The results reported here are steps along a path linking lipid signaling with epigenetic regulation.
Materials and Methods
Plant material, PtdIns(5)P and PtdIns(4)P treatments.
Arabidopsis Col 0 wild type seeds were sterilized and sown in 40 ml of Arabidopsis germination media (0.5X strength MS salts, 0.8% agar (w/v), 1.5% sucrose (w/v), pH 5.7, plus FeSO4 + NaEDTA and B5 vitamins), cold treated (4°C) for 48 hr and then seedlings were grown at 24°C under a cycle of 14 hr light/10 hr darkness. 1 mM stock solutions were made of D-myo-phosphatidylinositol 5-phosphate (PtdIns(5)P) and D-myo-phosphatidylinositol 4-phosphate (PtdIns(4)P) (Echelon Biosciences Inc.), following the manufacture's instructions. 10 µl of the stock were dissolved in 10 ml of the germination media without the agar (1 µM final concentration) and were added to the plants at specified stages of development. In experiments where the lipid-containing solutions were added to wild type plants at bolting, the majority of plants had initiated inflourescent stem development but stems were less than 2 cm high. Control samples were mock-treated with solutions not containing the lipid. Pictures and tissues collected at different times are as indicated.
RNA sample and microarray preparation.
In two independent replicate experiments, RNAs were isolated from PtdIns(5)P-treated, from PtdIns(4)P-treated, and from mock-treated control plants, grown and handled under the same conditions. Whole plants, grown for 20 hours in the presence of exogenously added drug, were harvested and quickly transferred into liquid nitrogen. Total RNA was extracted from frozen plants using TRIzol reagent following the manufacturer's instructions (Invitrogen) and further purified using Qiagen RNeasy column (Qiagen). Fifteen micrograms of total RNA was used to synthesize cDNA using Affymetrix One-Cycle cDNA Synthesis Kit according to the manufacturer's instructions (Affymetrix). All sample preparations followed prescribed protocols (Affymentrix Genechip Expression Analysis Technical manual). Hybridization was done on an Affymetrix Arabidopsis Genome ATH1 Array, stained with streptavidin-phycoerythrin conjugate on an Affymentrix Fluidics Station 450, followed by scanning with the GeneChip Scanner 3000 (Affymetrix). Affymetrix GeneChip Operating Software (GCOS) was used for washing, scanning, and basic data analysis.
Microarray data analyses.
The Affymetrix microarray contains more than 22,500 probe sets, representing approximately 24,000 genes. Each probe set consisted of 11 probe pairs with a perfect mach (PM) sequence corresponding to a specific region of a gene. For each PM sequence, there was also a corresponding mismatch (MM) oligo that differs by one base. In total, eight microarray hybridizations (including two control chips) were carried out and each experimental sample was analyzed versus each of the two wild type control sets. The data were first analyzed with Affymetrix GeneChip Operating Software (GCOS) that uses the MAS5 statistical algorithm, and then mined for significant genes with the AffyMiner program, a data mining tool developed by the Bioinformatics Core Research Facility of University of Nebraska-Lincoln (http://biocore.unl.edu/affyminer/). The data analysis by GCOS was performed in two steps: signal array analysis and comparison analysis. In signal array analysis, data were first processed for background correction, value extraction, data summarization and normalization. The One-Step Tukey's biweight method was used to estimate the variance among different probe pairs within a probe set and to produce a single expression index that represented transcript abundance. The One-Sided Wilcoxon signed-rank test was used to generate the Detection p-value and to make a Detection call as Present (P, p < 0.04) or Absent (A, p > 0.06). Array normalizations were preformed using the scaling approach, which adjusts the average intensity of every array to a common value in order to make the arrays comparable. The target signal intensity 500 was set up for scaling.
For comparison analysis, the array for wild type (wt) is designed as a baseline and the arrays for PtdIns(5)P- or PtdIns(4)P-treated samples were designed as treatment. Instead of simply comparing signal values of each probe set, MAS5 examines changes in the intensities of both PM and MM probes between the treatment and the baseline using the One-Sided Wilcoxon rank test to generate a Change p-value and make a Change call. The One-Step Tukey's biweight method was used to estimate the variance in ratios of probe pair intensities across the two arrays and to generate a single ratio that represented the magnitude and direction of change of a transcript expressed as Signal Log Ratio (base 2).
To compare two replicates for the treatment (e.g., PI5P1 and PI5P2) and two replicates for the control (i.e., wt01 and wt02), the data were published from GCOS to Affimetrix Data Mining Tool (DMT) for the calculation of average signal value and fold change for each probe set. The data exported from DMT were then analyzed with AffyMiner for identifying genes highly expressed in PtdIns(5)P-treated and PtdIns(4)P-treated samples when compared with wild type and to find overlapping genes that expressed significantly in PtdIns(5)P -and in PtdIns(4)P-treated samples. The following criteria were used by AffyMiner: (i) detection call should be “present” in the 2 experiment replicates; (ii) change calls from the pair-wise comparisons should be all “I”, i.e., increase, or ”D”, decrease; (iii) the fold change of average signal values between the treatments and the controls should be no less than 1.5. Five genes showing extreme values of fold change have been excluded from statistical analysis when determining the Pearson correlation coefficient.
The quality of replicate arrays was evaluated using scatter plots and pair-wise correlations. The scatter plot is a useful graphical tool for studying the spread and linearity of the data from two replicate arrays. When two arrays are perfectly linearly related, the points in the scatter plots fall on a straight line. The correlation of the two arrays represents the degree to which the two arrays are related. The cutoff for average signal log ratio between the control and experimental samples was 0.5. This corresponds to ∼1.5 fold change in intensity levels (amounts of expressed messages, respectively) between the experimental and control data sets.
RT-PCR analysis.
Total RNA was extracted from 0.3 g of tissue by using the BRL Trizol reagent and re-purified with the Qiagen RNeasy Mini Kit, following the manufacturers instructions. RT reactions were performed in a 20 µl volume containing 2.5 µg of total RNA and 200 units of the M-MLV Reverse Transcriptase from Invitrogen, following the manufacturer conditions. The TaKaRa Ex-Taq polymerase was used during the PCR reaction, and the conditions were as follow: 95°C for 30”, 60°C for 30” and 72°C for 1′ for 34 cycles. Specific gene primers were designed according to gene-specific sequences available at the Affimetrix web site.
Image analysis.
Images were captured with the Gel Doc 2000 gel documentation system and analyzed with the Quantity One software, both from Bio-Rad. To measure the amount of particular bands, a volume rectangle of the same size was created around each band of interest and the intensity data (the sum of intensities of the pixels inside the volume boundary area) were compared with the data of other objects using the Volume Analysis Report via the Global background subtraction method. The results are reported as differences in intensity (INT) or as percentage, when compared to the wild type samples.
Confocal-laser-scanning-microscopy.
Living roots, mock-untreated or exposed to the lipid were analyzed under an upright Leica TCS4D (488 nm line of a Kr/Ar laser) and an inverted Zeiss LSM 510 Meta microscopes. C-05R16 BODIPY-PI5P-tagged product from (Echelon Biosciences Inc., Salt Lake) was used to illustrate internalization and colocalization of exogenous PtdIns(5)P.
Results
Exogenously added PtdIns(5)P is taken up by Arabidopsis root cells.
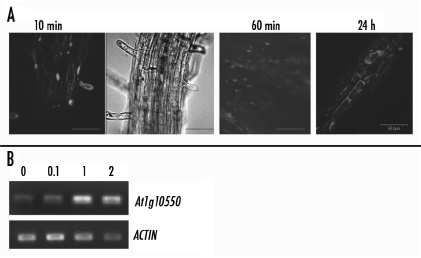
Apparently, the endogenous concentration of PtdIns(5)P is very low which may explain why this lipid has remained elusive. However, cellular PtdIns(5)P levels increased under osmotic stress4,5 consistant with a signaling function for this lipid. In order to explore effects triggered by elevated lipid levels, we supplied PtdIns(5)P exogenously, by adding it to the growth media or by submerging plant roots into liquid media containing the tested lipid compound (see individual experiments). Earlier, we have demonstrated that exogenoulsly added PtdIns(5)P co-localized with ATX1 inside cells and the two co-regulated a set of shared genes.20 Here, we show that a fluorescent red-tagged PtdIns(5)P added to the liquid media is uptaken initially by root hair cells and that, later, it may be massively present inside root cells (Fig. 1A). The signal was still present in cells 24 hours after treatment but it is unclear whether at this point the lipid was intact or whether the signal was associated with a metabolized product. Relevant for our further studies, however, is the fact that 20 hours after exposure to the lipid (the time-point when tissues were harvested for microarray hybridization), a set of Arabidopsis genes showed a reproducible robust change in expression (Fig. 1B). This result suggested that increased PtdIns(5)P levels triggered specific genome responses regardless of whether the ligand was still intact (see further below).
Figure 1.
Uptake of exogenously added PtdIns(5)P by root cells and expression of a target gene. (A) Uptake of PtdIns(5)P by Arabidopsis roots exposed to liquid media containing 1 µM PtdIns(5)P. Distribution of the tagged-(C-05R16 BODIPY-PI5P) lipid (red signal). Bars are 50.0 µm. (B) The expression of the At1g10550 gene (wall-modifying function) is tested 24 hours after exposure to different concentrations of PtdIns(5)P. Numbers on top represent concentrations in µM; The ACTIN7 gene was used as an expression and loading control.
To determine an optimal range of concentrations for our experiments, we tested the ability of PtdIns(5)P to alter gene-expression at three different concentrations. The expression of a PtdIns(5)P stimulated gene (At1g10550) is shown24 hour after exposure to 100 nM, 1 µM and 2 µM PtdIns(5)P (Fig. 1B). Because 0.1 µM PtdInsP caused bearly detectable differences, while 1 µM and 2 µM concentrations caused comparable differences, we have chosen to use 1 µM PtdInsP for our microarray and growth-testing experiments.
Genome-wide expression analyses.
ATH1 genome arrays were used to analyze genome-wide changes of Arabidopsis thaliana genes' expression in response to elevated levels of PtdIns(5)P. PtdIns(4)P was used as a control for the specificity. Affymetrix gene chips carrying 22,500 probes (∼24,000 Arabidopsis genes) were used for whole-genome expression analysis of plants treated with exogenously added 1 µ PtdIns(5)P or 1 µM PtdIns(4)P. To obtain a broader representation of genes affected by increased lipid levels, we chose a stage in development (bolting) when root, leaf and flower genes would be expressed. In two independent experiments, RNAs were isolated from control mock-treated, from PI5P-treated, and from PI4P-treated plants grown and handled under the same conditions. Expression patterns reflected whole-plant gene expression and not tissue-specific profiles.
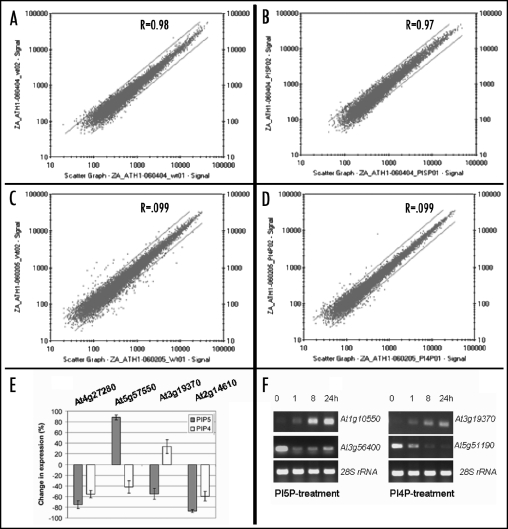
Experiments were performed in separate duplicate hybridizations with two independently isolated RNA samples. Data from each treated sample were analyzed versus each of the controls. Four separate independent preparations from wild type mock-treated plants were performed over a 10 month time span. Reproducibility of the four pairs of replicate arrays was illustrated by the scatter plots and confirmed by their associated correlation coefficients (Fig. 2A–D). The data show excellent agreement between the two arrays and have correlation coefficients in a range of 0.97 to 0.99 for data points with ‘present calls’ on both arrays. The numbers of total genes detected by eight independent hybridizations were: 60.4% and 57.4% for the wild type, 57.7% and 58.5% for the PtdIns(5)P-treated plants, and 60.7% and 62.2% for PtdIns(4)P-treated plants (correlation coefficients of 0.98, 0.97 and 0.99, respectively). In experiments performed 10 months later, the numbers of detected genes were 61.8% and 63.4% for control samples (correlation coefficient 0.99). The results indicate that our analyses consistently detected 60% (∼14,800) of all Arabidopsis genes supporting the validity of the detection technique. The majority (>99%) of significantly expressed genes showed less than two-fold variation in signal intensity between different RNA preparations. For mining significant genes and the criteria used to define genes with robust change in expression see Materials and Methods.
Among 14,800 detected genes expressed at this particular developmental stage, about 366 genes (∼3%) have responded to PtdIns(5)P by altering expression more than 1.5 fold (p << 0.05). Robust decrease in signal was found for 190; 176 showed increased expression signals. Changes in signal levels peaked at ∼10 fold (At3g28540 encoding an AAA-type ATPase) and ∼5 fold (At1g10550, encoding a xyloglucan endotransglycosylase) for the downregulated and the upregulated fractions, respectively. PtdIns(5)P activated and silenced genes are summarized in Supplemental Table 1 (Table ST1) online.
Table 1.
Association of PI5P- and PI4P-responding genes with cellular substructures: Classification according to their GOCC ID
| Downregulated genes | ||||||
| Endomembrane/wall genes | Chloroplast genes | Mitochondrian genes | Nucleus genes | Cytoplasm genes | Unspecifieda genes | Uknownb genes |
|---|---|---|---|---|---|---|
| PI5P (190 total) | ||||||
| 45 (24%) | 31 (16%) | 12 (6%) | 35 (18%) | 7 (4%) | 59 (32%) | 23 (12%) |
| PI4P (196 total) | ||||||
| 74 (38%) | 17 (9%) | 15 (7%) | 21 (11%) | 3 (1.5%) | 66 (33.5% ) | 21 (7%) |
| Upregulated genes | ||||||
| PI5P (176 total) | ||||||
| 84 (48%) | 15 (8%) | 12 (7%) | 9 (5%) | 6 (3%) | 50 (28%) | 14 (8%) |
| PI4P (95 total) | ||||||
| 30 (34%) | 12 (14%) | 9 (8%) | 3 (3%) | 8 (9%) | 28 (32%) | 25 (23%) |
Proteins with no assigned GOCC ID;
Expressed proteins with unknown predicted functions and no assigned GOCC ID. These entries are a subfraction of the group Unspecified;
*The three genes associated with nuclear functions do not include known Transcription Factors..
Approximately 291 genes (ů2% of all detected genes) showed significantly altered expression (p << 0.05) after treatment with PtdIns(4)P: 95 genes were up- and 196 genes were downregulated compared with mock-treated control samples (Table ST2 2, online). Change in signals peaked at ∼24 fold (expressed protein, At5g15420) and ∼21 fold (allergen family protein, At1g35310) in the down- and in the upregulated fractions, respectively.
To verify the microarray data, we performed RT-PCR on several shared genes altering expression in response to both PtdIns(5)P and PtdIns(4)P. The probes to be amplified were selected to illustrate cases of genes affected in the same, as well as in opposite, directions by the two lipids. RNAs from two different isolations were used as templates and the bands from each amplification reaction were quantified. The change in value from the control (wild type) was plotted on a histogram (Fig. 2E). The RT-PCR of tested genes showed changes in mRNA expression in general agreement with the microarray hybridization results.
Figure 2.
Reproducibility of GeneChip hybridization data. Scatter plots comparing the signal intensities of two independent replicate samples. (A) Mock-treated control sample; (B) PtdIns(5)P-treated sample; (C) Mock- treated control samples from an experiment performed 10 months later; (D) PtdIns(4)P-treated sample. Lines indicate 2-fold difference between replicates. (E) RT-PCR analysis confirming data from the microarray hybridizations. Samples were selected to represent changes in signal ratio, including subtler alterations, rather than those with the highest ratios in the microarrays. Differences in band intensities, after subtracting the values from wild type control samples, were plotted as percent change. Black columns represent changes in expression of PtdIns(5)P-treated samples and empty columns represent changed intensities for PtdIns(4)P-treated samples. The numbers on top are the gene TAIR ID numbers; (F) Levels of gene expression as a function of time after exposure to PtdIns(5)P and PtdIns(4)P. In three independent experiments, adult leaves were harvested from plants exposed to the respective lipid for the indicated periods of time (numbers on top of lanes indicate hours of exposure). RNAs were isolated and used as templates for RT-PCR amplifications of tested genes. Numbers on the right are TAIR ID numbers of tested genes. 28S rRNA is used as amplification and loading control.
PtdInsP-affected gene expression was stable at least 24 hours.
The kinetics of induction and maintaining of induced expression in target genes was examined at several time points. RNA was isolated from leaves of adult flowering plants before treatment and after 1, 8 and 24 hours exposure to PI5P or to PI4P. PT-PCR analyses showed that different genes displayed different time-responses (Fig. 2F). Relevant for our further studies is the fact that the samples taken for microarray hybridizations (after 20 hours exposure to the lipids) represented stabilized levels of altered gene expression.
Categorization of PtdIns(5)P- and PtdIns(4)P-affected genes according to function.
The distribution of the genes with significantly altered expression levels according to their functional characterization (based on the Genome Ontology, GO) indicated that PtdIns(5)P treatment has affected genes involved in diverse cellular and organismal processes (Suppl. Fig. 1, online). The largest proportion of impacted genes was involved in metabolic and physiological processes followed by genes involved in stimuli response, cell communications, and apoptosis. About 5% represented genes with regulatory functions, including epigenetic regulations, development and homeostasis.
PtdIns(4)P -affected genes encoded a similarly broad spectrum of functions, according to their GO-assignment (Suppl. Fig. 2, online). Notable differences caused by each of the lipids were that approximately 25% of PtdIns(4)P-affected genes encoded metabolic functions, while the respective fraction constituted 55% of PtdIns(5)P-affected genes. Genes encoding cellular and organismal physiological processes were affected more by PtdIns(4)P (∼45%) than by PtdIns(5)P (∼20%) of their respective overall impacted genes. It is interesting to note also the differences in gene fractions encoding cell communication (4.30% in PtdIns(4)P versus 9.2% in PtdIns(5)P) and in the regulation of gene expression (4.56% and 0.4% in PtdIns(4)P and PtdIns(5)P, respectively).
Thus, analyses at overall distribution levels revealed two important features of the PtdIns-triggered gene responses: both lipids influenced expression of genes encoding similar biological functions but the proportion of genes involved in a particular biological function was different for each lipid. This fact suggested that PtdIns(5)P and PtdIns(4)P participate in nonoverlapping mechanisms provoking specific responses in Arabidopsis. Subsequent analyses supported this conclusion (see below).
Products of affected genes categorized according to subcellular localization.
PtdIns(5)P and PtdIns(4)P-responding genes were classified further according to the subcellular localization of encoded products based on the assigned Gene Ontology Cellular Component (GOCC) numbers (Table 1).
Genes responding to PtdIns(5)P.
PtdIns(5)P-downregulated genes. In the PtdIns(5)P-downregulated fraction, 32% (∼60) of the genes did not have predicted association with cellular substructures; about 20 genes encoded expressed proteins with unknown function and unknown compartmentalization. Approximately 16% of the genes encoded chloroplast, 6% encoded mitochondrial, 4% encoded cytoplasm and 18% encoded nuclear components. Twenty-four percent encoded membrane-and wall-associated proteins. The majority of the latter activities represented ABC-type transporters, metal transporters, potassium-channel proteins, members of the cytochrome P450 family and various enzymes involved in sugar and lipid metabolism.
Thirty-five downregulated genes encoded established or putative transcription factors Notably absent among the downregulated fraction were members of the MADS-box family (see Supplemental Material, online).
Genes involving protein kinases, phosphatases and putative components of signal transduction pathways were also downregulated by PtdIns(5)P. Repressed were two Male Sterility (MS5) family genes (At5g48850, At1g04770) and two genes from the family of the Late Embryogenesis Abundant (LEA) encoding proteins (At1g52690, At5g06760) suggesting that PtdIns(5)P might impact the plant's reproductive, early developmental functions and responses to drought.
PtdIns(5)P-induced genes. One hundred seventy-six genes increased expression in response to the treatment. For the products of 50 genes (28%) there was no predicted subcellular location and, among them, 14 genes encoded expressed proteins with unknown function and unknown cellular compartmentalization. About 7% encoded mitochondrial, 8% encoded chloroplast, 3% encoded cytoplasm, 5% encoded nuclear and 48% encoded membrane and cell wall associated components. The latter represented a significant increase compared to the PtdIns(5)P silenced fraction. For all other organelle-associated PtdIns(5)P-responsive genes (except the mitochondrial), the percentage of upregulated genes was lower (about half) compared to the percentage of PtdIns(5)P-silenced genes. Activated were also genes involved in putative cellular transduction pathways, two-components signaling systems, protein kinases and phosphatases, genes for defense-response, disease resistance, heat shock, various stress-responses and apoptosis (for more analyses, see Supplemental Materials online)
Genes responding to PtdIns(4)P.
PtdIns(4)P-downregulated genes. After PtdIns(4)P treatment, fewer transcripts were detected for 196 Arabidopsis genes (Table 1 and Table ST2, online). A majority, 74 genes (38%) of the entire PtdIns(4)P downregulated fraction encoded membrane- or wall-associated proteins. These included members of the cytochrome P450 family, pathogen-response factors, various enzymes involved in sugar and lipid metabolism. For 66 genes (33%) of the downregulated sample there was no predicted association with cellular substructures and, among them, ∼7% encoded expressed proteins with unknown function and unknown compartmentalization. Approximately 9% of the genes encoded chloroplast, 7% encoded mitochondrial, 1.5% encoded cytoplasm and 11% encoded nuclear components. Sixteen genes (∼8%) in the downregulated sample encoded established or putative transcription factors including predominantly plant-specific factors of the AP2 (4 members) and the WRKY families (6 members). Genes encoding protein kinases, phosphatases and putative components of signal transduction pathways were downregulated by PtdIns(4)P as well.
PtdIns(4)P- induced genes. Following exposure to PtdIns(4)P, 95 genes increased expression (Table 1 and Table ST2, online). For the products of 28 genes, there was no predicted subcellular localization; twenty genes among them encoded expressed proteins with unknown function and unknown cellular compartmentalization. Eight percent encoded mitochondrial, 14% encoded chloroplast, 9% encoded cytoplasm, 3% encoded nuclear, and 34% encoded membrane and cell-wall associated components. Activated were genes involved in putative cellular transduction pathways, two-components signaling systems, protein kinases and phosphatases, genes for defense-response, disease resistance, heat shock, various stress-responses. Only three upregulated genes associated with the nucleus, none of them encoding a transcription factor.
Common targets of PtdIns(5)P and PtdIns(4)P networks: gene cluster analysis.
The data so far indicated that PtdIns(5)P and PtdIns(4)P influenced the expression of genes encoding similar functions in Arabidopsis, suggesting that the two messengers could use the same, or largely overlapping, pathways. To address the question of whether the plant discriminated between the two challengers and whether it used the same or distinct response mechanisms, we performed overlap analyses of PtdIns(5)P-and PtdIns(4)P-affected genes. Shared target genes would support the idea that PtdIns(5)P and PtdIns(4)P use common pathways to influence their expression.
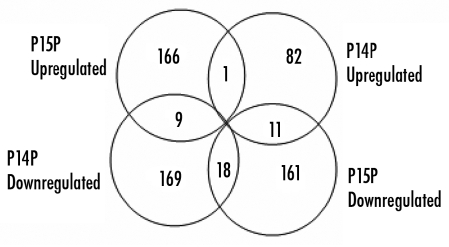
Gene cluster analyses were performed with PtdIns(5)P- and PtdIns(4)P-gene sets in all four possible combinations (see Methods). Overlapping genes are shown as Venn diagrams (Fig. 3) and are summarized in Table 2. Nineteen genes were co-regulated (18 down-, one upregulated). Twenty common genes changed expression in opposite directions: nine genes went up- after PtdIns(4)P, down-after PtdIns(5)P; eleven genes were repressed by PtdIns(4)P but activated by PtdIns(5)P. These results illustrate high selectivity of the gene-responses and are discussed in more detail below.
Figure 3.
Venn diagrams showing overlapping genes in PtdIns(5)P and PtdIns(4)P affected gene-sets. Overlapping genes with significantly altered expression from the microarray hybridization experiments were examined in all possible combinations. Same genes regulated in opposite directions by PtdIns(5)P and PtdIns(4)P underscore the selectivity of the response mechanisms.
Table 2.
Common set of Arabidopsis genes with significant changes in expression levels (cutoff 1.5 fold change) in response to treatment with PI4P and with PI5P
| Probe Set ID | Fold Change | Gene Title | AGI | Cellular Component | |
|---|---|---|---|---|---|
| PI5P/wt | PI4P/wt | (Gene Ontology ID) | |||
| Downregulated by both PI5P and PI4P | |||||
| 248448_at | −5.17 | −1.71 | AP2 domain-containing transcription factor, putative | AT5G51190 | |
| 249754_at | −2.36 | −1.92 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | AT5G24530 | |
| 253915_at | −3.14 | −1.72 | calcium-binding EF hand family protein | AT4G27280 | chloroplast (9507) |
| 254231_at | −2.20 | −1.95 | WRKY family transcription factor | AT4G23810 | |
| 254767_s_at | −1.76 | −1.68 | cytochrome P450 71A19, putative (CYP71A19) | AT4G13290 | endomembrane system (12505) |
| 254926_at | −2.02 | −1.78 | 1-aminocyclopropane-1-carboxylate synthase 6/ACC synthase 6 (ACS6) | AT4G11280 | |
| 251400_at | −2.07 | −3 | expressed protein | AT3G60420 | |
| 251705_at | −2.74 | −3.08 | WRKY family transcription factor | AT3G56400 | |
| 252193_at | −1.81 | −1.44 | myb family transcription factor | AT3G50060 | nucleus (5634) |
| 256245_at | −1.73 | −1.68 | heat shock protein 70, putative / HSP70, putative | AT3G12580 | |
| 263539_at | −2.79 | −1.89 | aminotransferase, putative | AT2G24850 | |
| 266385_at | −3.38 | −2.03 | pathogenesis-related protein 1 (PR-1) | AT2G14610 | eXTHacellular (5576), endomembrane system (12505) |
| 265837_at | −3.84 | −1.59 | expressed protein | AT2G14560 | |
| 263852_at | −2.53 | −2.31 | MutT/nudix family protein | AT2G04450 | |
| 261892_at | −2.25 | −1.55 | WRKY family transcription factor | AT1G80840 | |
| 245734_at | −1.60 | −1.69 | hydrolase, alpha/beta fold family protein | AT1G73480 | cytoplasm (5737), chloroplast (9507) |
| 264400_at | −2.33 | −2.07 | glucose-6-phosphate/phosphate translocator, putative | AT1G61800 | chloroplast (9507), membrane (16020), integral to membrane (16021) |
| 259561_at | −2.22 | −1.58 | wall-associated kinase 1 (WAK1) | AT1G21250 | eXTHacellular matrix (5578), plasma membrane (5886 |
| Upregulated by both PI5P and PI4P | |||||
| 264433_at | 1.77 | 2.35 | glycosyl hydrolase family 1 protein | AT1G61810 | endomembrane system (12505 |
| Downregulated by PI4P and upregulated by PI5P | |||||
| 247406_at | −1.74 | 1.86 | two-component responsive regulator/response regulator 6 (ARR6) xyloglucan:xyloglucosyl transferase/xyloglucan endotransglycosylase/ | AT5G62920 | |
| 247925_at | −1.51 | 2.18 | endo-xyloglucan transferase (TCH4) xyloglucan:xyloglucosyl transferase/xyloglucan endo- | AT5G57560 | cell wall (5618) |
| 247866_at | −1.80 | 3.56 | xyloglucan transferase (XTH3 | AT5G57550 | endomembrane system (12505) membrane (16020), integral to membrane |
| 249955_at | −2.02 | 1.53 | sugar transporter, putative | AT5G18840 | membrane (16021) endomembrane |
| 253667_at | −1.79 | 1.59 | peroxidase, putative two-component responsive regulator/response regulator 5 (ARR5)/ response reactor 2 | AT4G30170 | system (12505) |
| 252374_at | −1.50 | 2.32 | (RR2) | AT3G48100 | |
| 259879_at | −2.25 | 2.11 | calcium-binding EF hand family protein | AT1G76650 | chloroplast (9507) endomembrane |
| 245757_at | −1.85 | 4.5 | phosphate-responsive protein, putative | AT1G35140 | system (12505) |
| 266316_at | −2.65 | 1.92 | |||
| Upregulated by PI4P and downregulated by PI5P | |||||
| 248344_at | 1.71 | −2.37 | protein transport protein-related COP1-interactive protein 1/ | AT5G52280 | mitochondrion (5739) |
| 249271_at | 1.53 | −2.45 | CIP1 | AT5G41790 | cytoskeleton (5856) |
| XH/XS domain-containing | |||||
| 255566_s_at | 1.76 | −1.61 | protein | AT4G01780 | |
| 256746_at | 1.50 | −1.28 | glucan phosphorylase, putative | AT3G29320 | chloroplast (9507) |
| 258017_at | 1.79 | −1.84 | expressed protein | AT3G19370 | chloroplast (9507) |
| 258333_at | 1.55 | −2.01 | matrix-localized MAR DNA-binding protein-related | AT3G16000 | thylakoid membrane (9535), plastid nucleoid (42646) |
| 263296_at | 1.70 | −1.81 | calmodulin-binding protein-related | AT2G38800 | |
| 265464_at | 1.60 | −1.6 | myosin heavy chain-related | AT2G37080 | chloroplast (9507) |
| 259794_at | 1.58 | −1.87 | myosin heavy chain-related | AT1G64330 | |
| 261844_at | 1.55 | −2.12 | expressed protein | AT1G15940 | |
| 263112_at | 2.11 | −2.81 | kinase interacting family protein | AT1G03080 | |
Functionally related genes respond similarly, but selectively, to PtdIns(5)P.
Although PtdIns(5)P influenced expression of genes involved in a diversity of biological functions, some activities were remarkably overrepresented (Suppl. Fig. 1, online and Table 3). For example, 38% of the PtdIns(5)P-upregulated fraction represented simultaneously activated genes encoding key wall-architecture activities. The data suggested that a putative PtdIns(5)P-involving mechanism targeted selectively these genes and provided an opportunity to test the biological relevance of a PtdIns(5)P.
Table 3.
Cell wall modifying genes impacted by PI5P and PI4P
| ID | Gene ID | Increase/Decrease | ||
|---|---|---|---|---|
| PI5P | PI4P | |||
| XTH | 263598 | At2g01850 | 2.07 | |
| 263207 | At1g10550 | 4.59 | ||
| 257203 | At3g23730 | 2.59 | ||
| 255433 | At4g03210 | 1.7 | ||
| 254042 | At4g25810 | 2.5 | ||
| 253666 | At4g30270 | 2.26 | ||
| 251192 | At3g62720 | 1.85 | ||
| 247925 | At5g57560 | 2.18 | (−1.52) | |
| 247866 | At5g57550 | 3.56 | (−1.80) | |
| 247162 | At5g65730 | 1.6 | ||
| 245794 | At1g32170 | 1.66 | ||
| 245325 | At4g14130 | 2.48 | ||
| 266376 | At2g14620 | (−3.25) | ||
| 257102 | At3g25050 | - | 1.87 | |
| 256584 | At3g28750 | - | 2.84 | |
| Trans-Glucosylases# | 264857 | At1g24170 | 1.6 | |
| 264704 | At1g70090 | 2.97 | ||
| 260914 | At1g02640 | 3.12 | ||
| 255506 | At4g02130 | 1.82 | ||
| 248622 | At5g49360 | 3.16 | ||
| 253628 | At4g30280 | 3.46 | (−1.64) | |
| 253401 | At4g32870 | - | (−1.62) | |
| 251625 | At3g57260 | - | (−2.42) | |
| Arabinogalactans | 267260 | At2g23130 | 2.29 | |
| 265066 | At1g03870 | 2.56 | ||
| 259072 | At3g11700 | 1.63 | ||
| 250933 | At5g03170 | 2.36 | ||
| 249037 | At5g44130 | 2.44 | ||
| 248252 | At5g53250 | 3.33 | ||
| Expansins | 252997 | At4g38400 | 2.47 | |
| 252563 | At3g45970 | 3.66 | ||
| 266070 | At2g18660 | −2.21 | ||
| Pectin modifying | 254573 | At4g19420 | 2.31 | |
| 264931 | At1g60590 | (−2.36) | ||
| 260727 | At1g48100 | (−2.49) | ||
| 255524 | At4g02330 | - | (−1.73) | |
| 248714 | At5g48140 | - | 2.16 | |
| 258252 | At3g15720 | - | 1.97 | |
| 256966 | At3g13400 | - | 1.90 | |
| 258645 | At3g07850 | - | 8.0 | |
| 259269 | At3g01270 | - | 1.53 | |
| 259266 | At3g01240 | - | 2.43 | |
| 266764 | At2g47050 | - | 2.34 | |
| 256096 | At1g13650 | - | 1.96 | |
Downregulation of genes is shown in brackets.
Included are only members of families known to be involved in cell-wall metabolism.
Genes of the β-trans glycosyltransferases, xyloglucan endotransglucosylases (XET), xyloglucan hydrolases (XEH), expansins, arabinogalactans and pectin-modifying families encode cell wall-remodeling activities, involved in cell extension and, ultimately, in plant growth (reviewed in refs. 21–23). Of 36 cloned and putative members of the XET and XEH families in Arabidopsis collectively called, XTH (as suggested in ref. 24), 12 were up-, one downregulated, by PtdIns(5)P (Table 3). The response to PtdIns(4)P was remarkably distinct: only four genes were affected (two down-, two upregulated). The two PtdIns(4)P-downregulated genes were activated by PtdIns(5)P, while the two PtdIns(4)P upregulated genes responded only to PtdIns(4)P but not to PtdIns(5)P.
Six trans-glucosyltransferase family genes were stimulated by PtdIns(5)P; PtdIns(4)P silenced three. One gene, At4g30280, altered expression in opposite directions showing that PtdIns(4)P and PtdIns(5)P acted antagonistically upon this shared target. Genes belonging to the arabinogalactan and the expansin families were upregulated by PtdIns(5)P (only one expansin gene was downregulated), while no member of these families altered expression in response to PtdIns(4)P. In contrast, PtdIns(4)P stimulated expression of eight pectin-modifying factors and silenced one. PtdIns(5)P had a more limited effect upon these activities: one gene was up-, two downregulated (Table 3).
Thereby, PtdIns(5)P and PtdIns(5)P influenced non-overlapping sets of wall-modifying genes. The mechanisms engaging each of the PtdInsP are selective and highly specific. For instance, no gene from the group of the cellulose-synthase cell wall gene families (TAIR) was influenced by either PtdIns(5)P or PtdIns(4)P.
PtdIns(5)P and Arabidopsis growth.
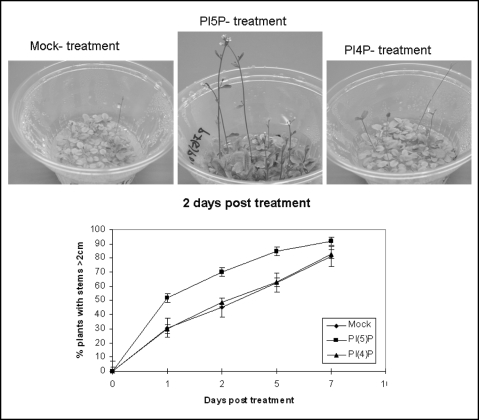
Cell-wall modifying activities underlie plant growth.21–26 The high representation of genes from these categories in the PtdIns(5)P-affected fractions suggested that PtdIns(5)P might influence growth in Arabidopsis. To test this idea, we measured stem elongation in PtdIns(5)P-treated plants. One µM PtdIns(5)P was added to plants at transition to bolting (9–10 rosette leaves). Stem length was measured after one, two, five and seven days. Differences in stem growth among PtdIns(5)P-treated and mock-treated samples were visible after one-day and were preserved up to seven days after exposure to the drug (Fig. 4; Table ST3, online). As plants grew older, the differences in stem growth became less pronounced and after seven days, most of the scored plants grew above 2 cm. Final lengths of mature plants were not measured.
Figure 4.
Effects of PtdIns(5)P and PtdIns(4)P on Arabidopsis stem growth. Arabidopsis plants were grown under the same growth conditions. At bolting stage, when most of the plants have developed stems smaller than 2 cm, the lipids were added to the growth media of experimental samples. Pictures show plants 2 days after exposure to 1 µM PtdIns(4)P (right-hand side panel), to 1 µM PtdIns(5)P (panel in the middle), and mock-treated with the solvent (left-hand side panel). The growth-curve in the bottom panel shows percentage of plants with stems longer than 2 cm measured at indicated days post treatment. Before treatment, all plants had stems shorter than 2 cm, reflected as an arbitrary point zero. Data are from three independent experiments (see Table 3, online for data from individual experiments). Bars show SD.
To establish whether stem growth was triggered by PtdIns(5)P, we have treated plants with PtdIns(4)P, under exactly the same conditions. The experiments were carried out in parallel, adding 1 µM PtdIns(4)P, instead of PtdIns(5)P. In contrast to PtdIns(5)P, PtdIns(4)P did not affect growth any differently than mock-treatment did (Fig. 4).
PtdIns(5)P-treated plants developed red stems and petioles, an effect not observed in the PtdIns(4)P-treated sample. These observations were consistent with microarray data indicating that PtdIns(5)P, but not PtdIns(4)P, affected pigmentation-related genes (Suppl. Figs. 1 and 2, online). These results provide arguments supporting the specificity of the genome responses and the validity of the microarray hybridization data.
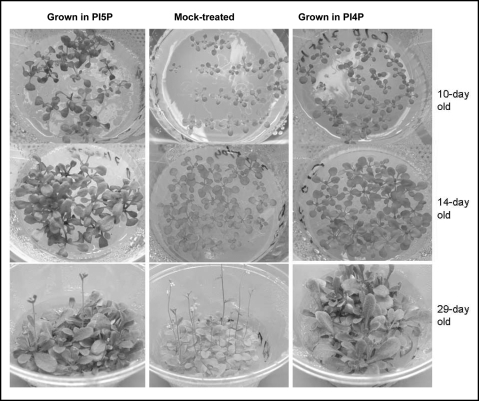
Presence of PtdIns(5)P and PtdIns(4)P early in development causes a different phenotype.
When present early in development, PtdInsP produced remarkably different phenotypes later in life. Seeds sown and grown in media containing 1 µM PtdIns(5)P developed faster than controls and at day 10, plants had 7–8 rosette leaves, bigger and greener than mock-treated controls (5–6 leaves). Accumulation of green mass in PtdIns(5)P-treated plants was even more pronounced in 14-day and in 29-day old plants (Fig. 5). Later, however, plants grown in PtdIns(5)P stayed longer in the vegetative state and developed inflorescence meristems 2–3 days later (13–15 rosette leaves) than control mock-treated plants (transition at 10 rosette leaves). The growth phenotypes of seeds sown and grown in the presence of PtdIns(4)P resembled the untreated sample early in development but bolted later than untreated and PtdIns(5)P-treated plants (18–20 rosette leaves at transition, Fig. 5). These results suggested that elevated PtdInsP levels early in development prolonged vegetative growth and stimulated accumulation of green mass.
Figure 5.
Effect of PtdIns(5)P and PtdIns(4)P on Arabidopsis plants sown and grown in PtdIns-containing media Control mock-treated seeds and plants are shown in the middle panel; samples sown and grown in PtdIns(5)P and PtdIns(4)P-containing media are shown on the left- and right-hand side, respectively. Pictures were taken under comparable magnification as seen by the rib-pattern of the plant containers.
Discussion
Microarray hybridization analysis revealed that approximately 14,800 Arabidopsis genes responded specifically to experimentally elevated PtdIns(5)P concentrations. High reproducibility across chips (correlation coefficients exceeding 0.95 for similar samples) supported the validity of our estimates. Exposure to PtdIns(5)P changed expression of ∼370 Arabidopsis genes. Comparable numbers of genes were silenced and induced (190 and 176, respectively) suggesting equally activating and repressive roles for PtdIns(5)P. In comparison, the analogue PtdIns(4)P, contributed to the altered expression of approximately 290 genes but more genes were down-(196) than upregulated (95). Many among the genes with altered expression are likely to be secondary targets reflecting the altered expression of a specific transcription factor gene by PtdIns(5)P or PtdIns(4)P.
Is PtdIns(5)P a signaling molecule?
The microarray data showing that both PtdIns(5)P and PtdIns(4)P influence gene activity suggested that the two molecules might be components of signaling mechanisms. The ability of phosphoinositides to penetrate hydrophilic and hydrophobic environments11 is compatible with the idea that they might act as signaling messengers.
Intracellular levels of PtdIns(5)P are low, in contrast to PtdIns(4)P, explaining why it has escaped identification for so long.5,6 Alternatively, PtdIns(5)P might be taken quickly by cell receptors or, be converted into bis-phosphate derivatives (PtdIns(4,5) P2 or PtdIns(3,5)P2. However, formation of PtdIns(5)P is considered to be a product of dephosphorylation of bis-phosphates.26,27 Increase in internal PtdIns(5)P levels upon exposure to 300 mM NaCl, suggested that PtdIns(5)P might function as a signaling molecule in osmotic stress.5,6
To establish whether PtdIns(5)P has a function on its own, we supplied it exogenously employing an adapted protocol.20 The possibility that addition of exogenous PtdIns(5)P might interfere with the balance of normal lipid concentration that this might provoke extreme or sporadic cell responses needs to be considered. However, it is important to point out, first, that in two independent experiments the same genes were found responding to elevated PtdIns(5)P or PtdIns(4)P, with correlation coefficients 0.97 and 0.99, respectively. These coefficients indicated high reproducibilty of detected respone-related genes and provided strong evidence that transcription responses were non random. Second, studies with exogenously supplied plant hormones and signaling molecules, i.e., salicylic acid, jasmonic aci, etc., resulted in breakthrough insights into the mechanisms of plant responses and associated pathways providing a precedent for the legitimacy of the approach here. Treating plants with exogenous PtdIns(5)P allowed us to identify the epigenetic factor ATX1 as an intracellular receptor for the ligand. The specific binding of PtdIns(5)P by ATX1, as well as the overlap of ATX1-PtdIns(5)P co-regulated genes, providing critical evidence that the two function in a novel signaling pathway.20 The abilty of ATX1 to bind selectively PtdIns(5)P, but not any of the related bis-phosphates, argued that PtdIns(5)P, itself, functions as a signaling molecule. It is important to note also that these results do not preclude a role of PtdIn(5)P as a precursor for the synthesis of derivatives or participation in cellular pathways not involving ATX1.
ATX1 did not bind phosphatidic acid, (D)-myo phosphatidylinositol, or diacylglacerol in protein-lipid-overlay assays, suggesting that fatty acid tails and non-phosphorylated head groups were not involved in the ATX1-interaction. Further, monophosphorylated PtdInsP other than PtdIns(5)P (i.e., PtdIns(3)P and PtdIns(4)P) as well as kinase products, i.e., PI4,5P2 and PI3,5P2 did not bind ATX1 as well, indicating that the head group phosphorylated at position 5 is the specific ATX1-interacting module (20). These facts argue that in the ATX1-mediated PtdIns(5)P response, the position of the phosphate on the head group determines the specificity of binding. Whether cellular lipases hydrolyze the uptaken PtdIns(5)P, leaving phosphoinositol 5-phosphate as the true ATX1-ligand, remains to be clarified.
It is important to note also that the signal observed in cells 24 hours after treatment does not indicate that the lipid has remained intact. Relevant for our analyses, however, is the fact that altered gene expression remained detectable for at least 24 hours (ST1 and ST2, online and Fig. 2E and F). These results suggest that either the compound remained stable and active throughout the tested period, or that its presence was not required to maintain changes that it had triggered initially. These fascinating possibilities merit further exploration.
PtdIns(5)P and PtdIns(4)P affect different genes and participate in distinct mechanisms.
PtdIns(5)P and PtdIns(4)P targeted the same families of transcription factors but affected different members within the families. Thus, 11 genes from the MYB family were found in the PtdIns(5)P target set, while PtdIns(4)P affected only one (At3g50060); PtdIns(4)P downregulated four genes from the plant-specific AP2 family, PtdIns(5)P affected one (At5g51190). PtdIns(4)P downregulated six WRKY family genes, PtdIns(5)P downregulated three. Five genes, among the 19 shared genes found in the PtdIns(5)P/PtdIns(4)P overlap, encoded transcription factors.
We found that the gene encoding PAT1, a protein preferentially binding PtdIns(5)P inside cells (28), was upregulated by PtdIns(5)P (Table ST1, online) suggesting a positive regulatory loop. Further, members within the PTL1 gene family were targeted specifically: only two among the six PATELLIN genes were affected by PtdIns(5)P, while none was affected by PtdIns(4)P.
PtdIns(4)P does not bind ATX1 and we propose that the molecular basis for the dramatically different effects caused by the position of one phosphate is the selectivity of their receptor proteins. This may correlate also with the distribution and functions of the two isoforms inside cells: PtdIns(4)P is membrane bound, while PtdIns(5)P has not been localized.
PtdIns(4)P and PtdIns(5)P affected genes belonging to families involved in response to other stimuli as well, like auxin, abscisic acid, gibberellin, cold, pathogens, etc. (refs. 29–31, and references therein). Our analyses revealed that different family members participate in each specific response (Table 2, Tables ST1 and ST2, online) suggesting distinct regulatory routs for each stimulus. However, different challengers could induce the same genes high-lighting points of convergence of distinct pathways. For example, the TCH4 gene (At5g57560) activated by brassinosteroids24 was activated also by PtdIns(5)P (Table 3). Some XTH gene family members raised their expression in response to IAA; other members of the same family did not change.24 The PtdIns(5)P-involving mechanism regulated a XTH gene-set distinct from IAA but overlapped with a IAA-pathway for two co-regulated genes (ref. 24; Table 3). Seven MYB genes were involved in sulfur-dependent response (14) and two among them (At1g56650 and At1g66390) correlated also with anthocyanin accumulation.32 The same two MYB genes (among the family of more than a hundred MYB genes) were also affected by PtdIns(5)P. This was consistent with increased pigmentation observed in the PtdIns(5)P-trated sample and with the microarray data (see also GO Distribution in Suppl. Fig. 1, online). In contrast, exposure to PtdIns(4)P did not cause a pigmentation phenotype or altered expression from pigmentation genes (Suppl. Fig. 2, online). Clearly, different members of the same family participate in separate networks providing specific responses to PtdIns(5)P or PtdIns(4)P. This fact indicates that the two messengers are involved in distinct pathways. Common gene targets would indicate points of convergence of the pathways; antagonistically responding genes indicate that the PtdIns(5)P and PtdIns(4)P pathways can counteract each other's effects.
The wall-modifying genes: a case for biological relevance of PtdIns(5)P-signaling.
Microarray hybridization analyses identified a remarkable group of functionally related (wall-modifying) genes that responded similarly to elevated PtdIns(5)P, but not to PtdIns(4)P. Concerted upregulation and selective responses to the two lipid isoforms underlined the specificity of the mechanism involving PtdIns(5)P and validated the biological relevance of this mechanism. An important link between two separate factors known to affect growth and development in Arabidopsis was revealed: on the one hand were the phosphoinositides, involved in pathways critical for cell survival, environmental adaptation and growth;1,7,11 on the other hand were the wall-modifying activities, recognized as forces regulating cell extension and plant growth (ref. 13 and refs. therein) whose expression was selectively influenced by PtdIns(5)P. The cell wall needs to loosen to allow expansion to occur in plant cells. This process is associated with modification of xyloglucan and the participation of enzymes cutting and rejoining the xyloglucan chains, in addition to activities including glucosylases, arabinogalactans, pectin-modifying enzymes and LTPs.21–23,33,34
An exciting question is whether differences in phenotypes triggered by PtdIns(5)P, but not by PtdIns(4)P at bolting, could be linked with the altered expression profiles of the wall-modifying genes. A smaller number of wall-modifying genes were affected by PtdIns(4)P than by PtdIns(5)P: PtdIns(5)P upregulated 26, downregulated two, genes from the XTH, trans-glucosylase, arabinogalactan and expansin families combined; PtdIns(4)P upregulated two and downregulated five members from this group. The results agree with a suggestion that each member of the wall-modifying families appears to be individually committed to a certain specific process that is regulated by different hormones.13 It is logical to expect that stem-elongation triggered by PtdIns(5)P reflected, at least in part, the altered expression patterns of the genes shown in Table 3. The upregulation of the majority of the PtdIns(5)P-affected wall-modifying genes may indicate a correlation between transcript levels of these genes and growing stems. In support, diminished transcript levels from the AtBXL1 gene (At5g49360) developed shorter stems under dry conditions.35 Exposure to PtdIns(5)P at bolting resulted in augmented transcription of AtBXL1 and development of longer stems (this study). PtdIns(5)P activated also a related gene, AtBXL2 (At1g02640); PtdIns(4)P did not activate any genes from this family, but repressed three. These results clearly implicate PtdIns(5)P in the regulation of Arabidopsis growth by modulating the expression of wall-modifying genes. However, it needs to be pointed out that it is not justified to draw a general correlation between increased transcripts levels from any wall-modifying family genes and cell growth. Wall-remodeling activities may both accelerate and suppress elongation of plant cells.36 Interactions among the various activities encoded by wall-modifying genes are complex requiring studies of individual members of each family.
Synthetic PtdIns5P and PtdIns4P might be stable for several weeks in the media solutions used here (personal communication, Dr. Paul Nilsen, Echelon). However, a possibility that the lipids might not be available to the end of the documented period (29 day-old plants) raises interesting issues of whether their presence was required for maintaining the effects and whether elevated lipids (even for a short period of time) at a certain developmental stage might have a long lasting effects displayed later in development. The phenotypes of 29 day-old plants differred dependent on whether they were exposed to the PtdInsP later in development or whether the PtdInsP were present at germination. This indicated that altered expression early in developement might cause phenotypic effects later in life. For example, the apparent overcrowding in the PtdInsP-treated samples compared to untreated controls (Fig. 5) resulted from the lipid-stimulated vegetative growth because same numbers of seeds were planted in the containers. Overcrowding was not so obvious when the PtdInsP were introduced to mature plants (Figure 4). Our data suggested that presence of PtdIns(5)P or PtdIns(4)P early in development stimulated production of leaf (vegetative) tissue but delayed stem growth. Addition of PtdIns(5)P, but not PtdIns(4)P, later in development stimulated stem elongation. It will be particularly interesting to compare microarray expression profiles of genomes exposed to PtdInsP at different developmental stages.
Collectively, our results suggest that plants respond specifically to augmented PtdIns(5)P and PtdIns(4)P levels reflecting their involvement in distinct mechanisms. Different members of the same families altered expression in response to PtdIns(4)P and PtdIns(5)P indicating that individual genes of a family were under selective regulation. The same genes may respond cooperatively, or antagonistically, to a variety of elicitors indicating that individual genes within a family are under the control of multiple factors. This might ensure highly specific responses to developmental and environmental cues.
Acknowledgements
This study was supported in part by NSF grant MCB-0343934 to Z.A.
Note
Supplemental material can be found at http://www.landesbioscience.com/journals/psb/supplement/alvarezvenegasPSB1-3-sup.pdf
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=2997
References
- 1.Wang X. Lipid signaling. Curr Opin Plant Biol. 2004;7:329–336. doi: 10.1016/j.pbi.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Meijer HJG, Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- 3.Charron D, Pingret JL, Chabaud M, Journet EP, Barker DG. Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 2004;136:3582–3593. doi: 10.1104/pp.104.051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Mitchell RH. Osmotic stress activates phosphatidyl-D-inositol 3,5-bisphosphate synthesis in yeast. Nature. 1997;190:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 5.Meijer HJG, Berrier CP, Iurisci C, Divecha N, Musgrave A, Munnik T. Hyperosmotic stress induces rapid synthesis of phosphatidyl-D-inositol 3,5-bisphosphate in plant cells. Biochem J. 2001;360:491–498. doi: 10.1042/0264-6021:3600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pical C, Westergren T, Dove SK, Larsson, Sommarin M. Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol phosphate and phosphatydylcholine in Arabidopsis thaliana cells. J Biol Chem. 1999;274:38232–38240. doi: 10.1074/jbc.274.53.38232. [DOI] [PubMed] [Google Scholar]
- 7.Jones DR, Divecha N. Linking lipids to chromatin. Curr Opin Genet Develop. 2004;14:196–202. doi: 10.1016/j.gde.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Roth MG. Lipid regulators of membrane trafficking through the Golgy complex. Trends Cell Biol. 1999;9:174–179. doi: 10.1016/s0962-8924(99)01535-4. [DOI] [PubMed] [Google Scholar]
- 9.Sbrissa D, Ikonomov O, Deeb R, Shisheva A. Phosphatidylinositol 5-phosphate biosynthesis is linked to PIKfyve and is involved in osmotic response pathway in mammalian cells. J Biol Chem. 2002;277:47276–47284. doi: 10.1074/jbc.M207576200. [DOI] [PubMed] [Google Scholar]
- 10.Tronchere H, Laporte J, Pendaries C, Chaussade C, Liaubet L, Pirola L, Mandel JL, Payrastre B. Production of phosphatidylinositol 5-phosphate by the phosphoinositide 3phosphatase myotubularin in mammalian cells. J Biol Chem. 2004;279:7304–7312. doi: 10.1074/jbc.M311071200. [DOI] [PubMed] [Google Scholar]
- 11.Stevenson JM, Perera IY, Heilmann I, Person S, Boss W. Inositol signaling and plant growth. Trends Plant Sci. 2000;5:252–258. doi: 10.1016/s1360-1385(00)01652-6. [DOI] [PubMed] [Google Scholar]
- 12.Jung J-Y, Kim Y-W, Kwak JM, Hwang J-U, Young J, Schroeder J, Hwang I, Lee Y. Phosphatidylinisitiol 3-and 4-phsphate are required for normal stomatal movements. Plant Cell. 2002;14:2399–2412. doi: 10.1105/tpc.004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minorsky PV. The wall becomes surmountable. Plant Phys. 2002;128:345–353. doi: 10.1104/pp.900022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of bosyntheitic pathways provides response specificity. Plant J. 2003;33:633–650. doi: 10.1046/j.1365-313x.2003.01657.x. [DOI] [PubMed] [Google Scholar]
- 15.Irvine RF. Nuclear lipid signaling. Nature Rev. 2003;4:1–12. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- 16.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinisitide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 17.Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 18.Steger D, Haswell ES, Miller AL, Wente SR, O'Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, An Arabidopsis homolog of Trithorax has histone methylase activity and activates flower homeotic genes. Curr Biol. 2003;13:627–637. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez-Venegas R, Sadder M, Hlavacka A, Balu_ka F, Xia Y, Lu G, et al. The Arabidopsis Homolog of Trithorax, ATX1, binds phosphoinositide 5-phosphate and the two regulate a common set of target genes. Proc Natl Acad Sci. 2006:6049–6054. doi: 10.1073/pnas.0600944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knox P. Intriguing, complex and everywhere getting to grips with arabinogalactan-proteins. Trends Plant Sci. 1999;4:123–125. [Google Scholar]
- 22.Rose JKC, Saladie M, Catala C. The plot thickens: new perspectives of primary wall modification. Curr Opin Plant Biol. 2004;7:296–301. doi: 10.1016/j.pbi.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Scheible WR, Pauly M. Glycosyltransferases and cell wall biosyntheiss: novel players and insights. Curr Opin Plant Biol. 2004;7:285–295. doi: 10.1016/j.pbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Xu W, Campbell P, Vargheese AK, Braam J. The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J. 1996;9:879889. doi: 10.1046/j.1365-313x.1996.9060879.x. [DOI] [PubMed] [Google Scholar]
- 25.Baluska F, Samaj J, Wojtaszek P, Volkman D, Menzel D. Cytoskeleton-plasma membrane- cell wall continuum in plants. Emerging links revisited. Plant Phys. 2003;133:482–491. doi: 10.1104/pp.103.027250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinchliffe KA, Ciruela A, Irvine RF. PIPkins, heir substrates and their products: new functions for old enzymes. Biochim Biophys Acta. 1998;1436:87–104. doi: 10.1016/s0005-2760(98)00140-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhong R, Burk DH, Morrison HWIII, Ye ZH. FRAGILE FIBER3, an Arabidopsis gene Encoding a Type II Inositol Polyphosphate 5-Phosphatase, Is Required for Secondary Wall synthesis and actin organization in fiber cells. Plant Cell. 2004;16:3242–3259. doi: 10.1105/tpc.104.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterman TK, Ohol YM, McReynolds LJ, Luna EJ. Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 2004;136:3080–3094. doi: 10.1104/pp.104.045369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Wang C, Sang Y, Qin C, Welti R. Networking of phospholipases in plant signal transduction. Plant Physiol. 2002;114:331–335. doi: 10.1034/j.1399-3054.2002.1150301.x. [DOI] [PubMed] [Google Scholar]
- 31.Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought and salt stress. Plant Cell. 2002;S:165–183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee Y, Dongsu C, Kende H. Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol. 2001;4:527–532. doi: 10.1016/s1369-5266(00)00211-9. [DOI] [PubMed] [Google Scholar]
- 34.Minic Z, Rihouey C, Do C, Lerouge P, Jouanin L. Purification and characterization of enzymes exhibiting _-D-xylosidase activities in stem tissues of Arabidopsis. Plant Phys. 2003;135:867–878. doi: 10.1104/pp.104.041269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goujon T, Minic Z, Amrani A, Lerouxel O, Aletti E, Lapierre C, Joseleau JP, Jouanin L. AtBXL1, a novel higer plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development. Plant J. 2003;33:677–690. doi: 10.1046/j.1365-313x.2003.01654.x. [DOI] [PubMed] [Google Scholar]
- 36.Takeda T, Furuta Y, Awano T, Mitsubishi Y, Hayashi T. Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci. 2002;99:9055–9060. doi: 10.1073/pnas.132080299. [DOI] [PMC free article] [PubMed] [Google Scholar]