Abstract
Cell migration is essential in many physiological and pathological processes. To understand this complex behavior, researchers have turned to quantitative, in vitro, image-based measurements to dissect the steps of cellular motility. With the rise of automated microscopy, the bottleneck in these approaches is no longer data acquisition, but data analysis. Using time-lapse microscopy and computer-assisted image analysis, we have developed a novel, quantitative assay that extracts a multivariate profile for cellular motility. This technique measures three dynamic parameters per single cell: speed, surface area, and an index of dynamic cell expansion/contraction activity (DECCA). Our assay can be used in combination with a variety of extracellular matrix components, or other soluble agents, to analyze the effects of the microenvironment on cellular migration dynamics in vitro. Our application was developed and tested using A431 and HT-1080 cell lines plated on laminin-332 or fibronectin substrates. Our results indicate that HT-1080 cells migrate faster, have a greater surface area, and have a higher DECCA index than A431 cells on both matrices (for all parameters, p < 0.05). Spearman's correlation coefficients suggest that for these cell lines and matrices, various combinations of the three measurements display low to medium-high levels of correlation. These findings compare well with previous literature. Our approach provides new tools to measure cellular migration dynamics and address questions on the relationship between cell motility and the microenvironment, using only common microscopy techniques, accessible image analysis applications, and a basic desktop computer for image processing.
Key words: migration, motility, live-cell imaging, microscopy, extracellular matrix, laminin, fibronectin, computer-assisted
Introduction
Cell migration is essential for many physiological and pathological processes such as normal embryonic development, inflammation, wound healing, angiogenesis and cancer metastasis.1,2 The process of eukaryotic cell migration includes a set of underlying, interlinked sub-processes including cytoskeletal reorganization, cell protrusion, attachment at the leading edge, cell contraction for physical translocation, and detachment of adhesion at the trailing edge of the cell.2–4 Furthermore, migration is a highly complex, dynamic process that is stringently regulated not only by internal cellular signals, but also by external cues from the surrounding microenvironment.5,6 Since cell migration is a process involving multi-component structures that are regulated in both space and time,2 it is necessary to employ experimental procedures that are capable of capturing the dynamics of cellular movement, in order to reveal information about the underlying mechanisms.4,7
Analysis of Dynamic Cell Size and Morphological Changes
Migration of epithelial cells follows a number of specific processes that are typically associated with specific changes in cell size and shape. Thus, one can gain insight into the specific mechanisms of cell movement by studying these morphological changes. Kymography is one method that has been used to gain insight into the mechanisms of actin, cortactin, and various other molecules involvement in membrane protrusion.8,9 This technique involves high-resolution time lapse microscopy to capture subcellular motion. Kymography is used for relatively small sample sizes (due to highly magnified imaging), during relatively short periods of time, for the study of subtle events such as actin polymerization at the leading edge.9–11 A number of methods have also been used to quantify morphological changes at the cellular level, including a diagonal measurement of cell elongation,12 human interpretation of shapes,13 and most commonly surface area measurements.14–18 These studies have looked at morphology as an indicator of differentiation, apoptosis, and various other processes.19,20 Traditional surface area measurements tend to rely on two types of analyses: edge detection through thresholding and edge-based segmentation.15 Some weaknesses of these techniques include the need for fluorescent cellular markers and an assumption of a particular cell shape (e.g., a round cell)—both of which can lead to complications, depending on your model system.15,21,22 For these reasons, surface area measurements must be optimized for specific cell types.15 Many epithelial cell lines spread out and lie flat against the surface they are seeded on, and have a wide variety of shapes, with multiple leading edges at once; this variety of shapes and decrease in contrast makes edge detection, by traditional means, much less accurate.15 Although a number of assays currently exist to examine cell morphology, both at the cellular and sub-cellular levels, there is still certainly room for new, quantitative techniques that can be used for accurate analyses of cell shape and other motility parameters.
Classical Motility/Migration Assays
The process of eukaryotic cell migration in various microenvironments has also been quantitatively measured using a variety of in vitro methods.4,7,23–26 These methods generally fall into two major categories: the first includes techniques that monitor the average behavior of a large populations of cells, such as classical wound-healing assays and Boyden chamber techniques;25 the second involves tracking individual cell motility, which commonly uses time-lapse video-microscopy.7,25,27 Both types of assays have their merits and limitations. Studying population motility allows researchers to obtain quantitative data very quickly, often with minimal expense, but only provides information about the average behavior of cells, often with minimal resolution. In contrast, single cell analysis allows the acquisition of more detailed information about cell motility (e.g., turn angle, velocity, cellular persistence), but the reaping of quantitative data can be extremely labor intensive. This second category of assays can also be designed to reveal subcellular dynamics of single cells, such as changes in cytoskeletal organization, lamellipodial protrusion and focal adhesion turnover.28,29
Single cell analysis is useful because it can be used to eliminate a number of variables that intrinsically influence classical wound-healing and Boyden chamber assays, such as cell-cell adhesion and proliferation.30–32 Furthermore, cells freshly removed from culture can be plated just prior to performing an experiment; whereas in wound-healing or Electric Cell-substrate Impedance Sensing (ECIS™) assays, cells must first be grown to confluence.25,33,34 Seeding cells just prior to time-lapse microscopy allows an experimenter to control the extracellular matrix (ECM) components, important variables in migration studies. ECM molecules, including proteins such as laminin (Ln) and fibronectin (Fn), play a critical role in regulating cell shape, polarity and growth.35,36 In classical assays, which often involve incubating cells for extended periods of time, experimenters are commonly burdened by cells laying down their own matrix components. This problem is further confounded in some experiments that capture population dynamics, as some investigators ignore this issue and may come to inappropriate conclusions. Although single cell analysis can potentially overcome many problems associated with these types of assays, it continues to be a labor-intensive measure of cell migration, due to its capacity for generating an abundance of quantitative data. In order to measure these parameters in a more efficient manner, development of computational methods for automation of data analysis is crucial.37,38
Computational Methods of Quantitation
With the addition of microscopes with automated stages, and the massive increases in processing power and data storage, it is now possible to produce enormous amounts of image data in an automated, or semi-automated, fashion. The bottleneck in imaging research is no longer data acquisition; it is now data analysis.39 Using computer-aided image analysis, or a semi-automated quantitative approach, is an ideal way to efficiently cope with complex processes such as cell behavior.37,40 Many researchers have utilized computer-assisted analysis, but semi- and fully-automated systems are still beyond most cell motility laboratories. A whole range of commercially available software programs have attempted to provide cell tracking software (e.g., MetaMorph, OpenLab and ImageJ plug-ins).41 These programs typically rely on thresholding, edge detection filters, or template matching—functions useful for capturing cell motility when applied appropriately. Some semi- and fully automated programs designed for analysis of amoebae, progenitor cells, and various other motile cell populations have also seen some success;19,37,38 however, such software has largely failed to catch on with eukaryotic cell migration researchers, due to high error rates associated with tracking these cells in phase contrast images.39 Manual tracking of cell speed currently remains the gold standard in the field.41 Other types of image analysis, such as kymography techniques used to determine cell shape changes, currently have no automation beyond stacking images through time, along a one-dimensional line.42 Here, we present custom algorithms that determine dynamic cell attributes, while overcoming many of the aforementioned issues with currently available techniques, and can be run on a typical computer found in almost any laboratory using only MetaMorph, MATLAB® and Microsoft Excel. In addition, we obtain a multivariate profile of individual cells that incorporates speed, surface area, and a third parameter of cell activity and shape change, which we refer to herein as Dynamic Expansion/Contraction of Cell Area (DECCA). An overview of our entire method for analyses is outlined in Figure 1.
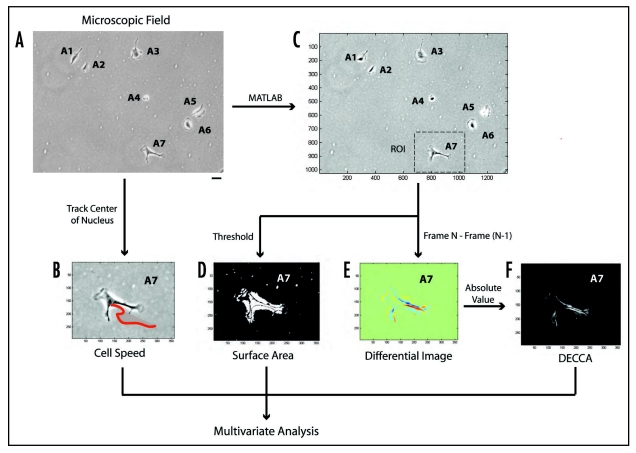
Figure 1.
Overview of Multivariate Profiling of Single Cells. This flowchart represents the step-wise progression of our image analysis technique, including: (A) phase-contrast image capture (six random fields per well, in duplicate) and manual application of unique identifying numbers to all cells (i.e., A1–A7); (B) tracking the center of each cell nucleus manually using Metamorph software to quantify cell speed; the applied red line represents cell tracks over the course of a movie. (C) selection of regions of interest (ROI) manually from original phase contrast images using MATLAB; (D) creating computer-generated thresholded images in MATLAB to calculate cell surface area, and (E) creating computer-generated differential images in MATLAB by subtracting the pixel intensities from one frame to the next. (F) Differential images were further processed by taking the absolute value of the pixel intensities to obtain the DECCA measurement. The scale bar seen in part (A) is equal to 100 µm. Image axes in (B–F) are MATLAB generated coordinates for each image and ROI.
Results
Cell speed quantitation.
A431 cells were seeded in microplates coated with either 1 µg/mL Ln-332 or 10 µg/mL Fn, and time-lapse microscopy experiments were performed, in duplicate, as described in Material and Methods. As displayed in Figure 2A, A431 cells exhibited a mean cell speed of 0.82 ± 0.44 µm/min on Ln-332 [grey; N = 3 (415 cells)] and 0.41 ± 0.21 µm/min on Fn [white; N = 2 (272 cells)]. Cell speed on Ln-332 substrate was found to be significantly faster than on Fn (p < 0.001). Furthermore, there was no significant difference (p > 0.05) between A431 cell speed measurements within duplicates performed each day, nor between repeated experiments, demonstrating the repeatability of our manual cell tracking method (results not shown).
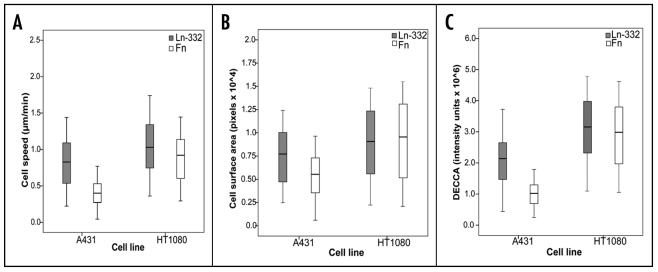
Figure 2.
Quantitation of Cell Speed, Surface Area and DECCA. A431 or HT-1080 cells were allowed to adhere to laminin-332 (Ln-332) or fibronectin (Fn) coated microplates for 1 h at RT. Time-lapse microscopy was used to capture cell motility for 4 h. (A) For cell speed quantification, cells' paths were tracked manually using MetaMorph software. On Ln-332 (grey), A431 (N = 415 cells) and HT-1080 (N = 254 cells) speed was calculated to be 0.82 ± 0.44 and 1.07 ± 0.44, respectively, which was found to be significantly different from one another (p < 0.05). Similarly, on Fn (white), A431 (N = 272 cells) and HT-1080 (N = 84 cells) speed was 0.41 ± 0.21 and 0.91 ± 0.37, respectively, which was also found to be significantly different from one another (p < 0.05). Each cell lines' behavior on the two substrates was also significantly different (p < 0.05). (B) Cell surface area measurements were captured using custom-written MATLAB algorithms, which removed background pixels via thresholding. All remaining pixels were taken to represent the cell. On Ln-332 (grey), A431 and HT-1080 cell surface area measurements were calculated to 7.762 ± 4.585 and 9.390 ± 5.092, respectively (p < 0.05); on Fn (white), measurements were calculated to be 5.878 ± 3.220 and 9,907 ± 5,102 (p < 0.05). (C). DECCA measurements were obtained using custom-written MATLAB algorithms, which took the absolute value of subtracted pixel intensities frame to frame, to produce a cell activity index. The DECCA of A431 cells on Ln-332 (grey) and Fn (white) was calculated to be 2.22 × 106 ± 1.33 × 106 and 1.05 × 106 ± 0.45 × 106 intensity units, respectively. The DECCA of HT-1080 cells on Ln-332 (grey) and Fn (white) was 3.20 × 106 ± 1.35 × 106 and 3.04 × 106 ± 1.33 × 106 intensity units, respectively. There was a significant difference between cell lines, with HT-1080 cells having a higher DECCA on both matrices (p < 0.001). There was also a significant difference on A431 DECCA on different matrices (p < 0.001), however there was no difference between matrices for HT-1080 cells (p > 0.05). All plots represent mean (—), 95% confidence interval (box), and standard deviations (whiskers).
HT-1080 cell speed was also examined by seeding cells either on 1 µg/mL Ln-332 or 10 µg/mL Fn-coated microplates, and experiments were performed, in duplicate, as above. As depicted in Figure 2A, the mean speed of HT-1080 cells was calculated to be 1.07 ± 0.45 µm/min on Ln-332 [grey; N = 2 (254 cells)] and 0.91 ± 0.37 µm/min on Fn substrate [white; N = 1 (84 cells)], which was found to be a significant difference (p < 0.05). Furthermore, there was no statistical difference (p > 0.05) found between duplicates, or across days of experimentation, further demonstrating the repeatability of our manual cell tracking results. In summary, both A431 and HT-1080 cells showed a reproducible and significant difference in mean cell speed, with HT-1080 cells migrating faster than A431 cells across both matrices (p < 0.05). In addition, both cell lines migrated faster on Ln-332 than on Fn (p < 0.001).
Surface area quantitation.
The same movies used to quantify cell speed in the previous results were also used to determine cell surface area measurements (as described at length in the Materials and Methods section). A blinded investigator produced this data, with no knowledge of cell speed results. Of all the cells captured during time-lapse microscopy for initial cell speed analysis, only 3.1% (34/1080 total cells) were lost during this surface area analysis, due to unfocused images or cells leaving frames mid-movie.
As depicted in Figure 2B, the mean cell surface area of A431 cells on Ln-332 and Fn substrates was calculated to be 0.78 × 104 ± 0.46 × 104 and 0.59 × 104 ± 0.32 × 104 pixels, respectively. Mean cell surface area of HT-1080 cells on Ln-332 (grey) and Fn (white) was 0.94 × 104 ± 0.51 × 104 and 0.99 × 104 ± 0.51 × 104, respectively. There was a significant difference between surface area measurements of the two cell lines, with HT-1080 cells having a higher surface area on both matrices (p < 0.001). This computer-assisted analysis (that HT-1080 cells were larger than A431 cells) was confirmed by the researcher who performed the manual cell tracking data analysis, demonstrating the accuracy of the computed-assisted surface area measurements. There was also a significant difference on A431 cell surface area on different matrices (p < 0.001), however, there was no difference between matrices for HT-1080 cells (p > 0.05).
DECCA quantitation.
The same movies used to quantify cell speed and surface area were also used to determine DECCA values (see Fig. 1 for clarification) for the analysis of total cellular motion (as described at length in the Discussion and Materials and Methods sections). Again, a blinded researcher produced this data, with no knowledge of cell speed results. The DECCA of A431 cells on Ln-332 and Fn was calculated to be 2.22 × 106 ± 1.33 × 106 intensity units and 1.05 × 106 ± 0.45 × 106 intensity units, respectively (Fig. 2C). The DECCA of HT-1080 cells on Ln-332 and Fn was 3.20 × 106 ± 1.35 × 106 intensity units and 3.04 × 106 ± 1.33 × 106 intensity units, respectively (Fig. 5B). There was a significant difference between cell lines, with HT-1080 cells having a higher DECCA on both matrices (p < 0.001). There was also a significant difference on A431 DECCA on different matrices (p < 0.001), however there was no difference between matrices for HT-1080 cells (p > 0.05).
Figure 5.
Dynamic Expansion/Contraction (DECCA) Algorithms for Image Analysis. (A) Custom-written, MATLAB algorithms were developed to measure DECCA. Blue text indicates an integrated MATLAB function, purple text indicates a string, and black is used for all other text and commands. (B) Sample phase contrast images of an HT-1080 cell (top row) and corresponding micrographs of differential (middle row) and DECCA images (bottom row) are presented for a sample motile cell over a short time series (205–220 min), for reference. In differential images, color is representative of the change in intensity between phase contrast images from frame to frame (blue is a negative intensity change, green is no change, and yellow/red is positive change). DECCA images are obtained by obtaining the absolute value of pixels present in differential images, and this value is averaged across an entire movie to produce a cell's DECCA measurement. The solid scale bar represents 100 µm.
Multivariate correlations.
Cell speed, surface area and DECCA measurements correlated significantly for all pairs, in both A431 and HT-1080 cell lines, and on both matrices (Fig. 3). Spearman's correlation coefficients (ρ) were calculated for the data grouped by cell line, matrix, and in its entirety. Speed vs. surface area typically showed a low correlation (ρ = 0.163–0.349), while speed vs. DECCA (ρ = 0.394–0.594) and surface area vs. DECCA (ρ = .434–0.545) both showed a medium level of correlation (Fig. 3).43 This trend indicates that larger cells tended to migrate faster, and have a higher cell activity level than smaller cells, as anticipated for these particular cell types. These results again demonstrate the validity of our method. Furthermore, since the trend was present in two cell lines, and on two different matrices, our method has been demonstrated to be reproducible for a variety of experimental conditions.
Figure 3.
Correlation Between Variables. Regression plots for A431 cells (A) and HT-1080 (B) cells on both Ln-332 (grey markers) and Fn (black markers). All Spearman's correlation coefficients (r) demonstrated various positive levels of correlation between the three measurements when grouping by both cell line and ECM substrate.
Discussion
Although our understanding of individual processes underlying cell migration continues to increase, major gaps in information concerning how they are coordinated spatially and temporally still remain.2 New techniques need to be developed that can bring insight into how these individual processes interact by quantifying dynamic cell movements and analyzing single cells in an automated manner. Computer-assisted, quantitative analysis of migrating cells provides an objective means of comparing migration properties of cells and yields insight into the underlying mechanisms of cell motility. Here, we report a dynamic multivariate analysis of single-cell motility that includes a combination of both novel algorithms/image analysis methods (surface area; DECCA) and existing techniques (cell speed). The first measurement we present, cell speed, was captured using a standard manual cell-tracking technique (Metamorph) from movies generated by time-lapse, phase contrast microscopy. The second measurement utilized a custom-written MATLAB algorithm designed to threshold images to calculate cell surface area. Obtaining surface area measurements gives us insight into the shape and overall health of cells. For example, epithelial cells tend to decrease their surface area when unhealthy or stressed and many cell lines change their shape upon differentiation, which is often reflected by a change in surface area. The third measurement, DECCA, is a novel measurement of total cell activity. That is, DECCA captures the pixel intensity change from one frame to the next, and averages these changes over the length of the movie. Thus, a higher DECCA value represents a higher amount of cell activity. This measurement is not necessarily a measurement of cell migration, as membrane protrusion without translocation, can also lead to high DECCA values. These three values were then combined using a unique identifier system to obtain three measurements per cell (for 1000+ individual cells, from 2 cells lines, on 2 substrates).
Our experimental results demonstrate the repeatability and reliability of our technique. Our data indicate low to medium-high correlations between all three measurements, depending on the particular cell line and substrate combinations. This range of relationships was anticipated for these cells, due in part to their high migration rates, and various shape changes observed in culture, when plated on Ln-332 or Fn.44,45 However, given different experimental guidelines (i.e., cell lines, ECM components, or introduction of mutations), these trends may certainly change. For example, NRK49F cells with defects in Rho or adducin have been shown to have active lamellipodial ruffling, while being unable to migrate.46 Based on these findings, we hypothesize that these mutant cells would have an unchanged cell surface area and DECCA (compared to wild type), but their cell speed would decrease drastically. Furthermore, inclusion of leukocytes, or various other immune cells, may also significantly alter results, as these cells are commonly very motile, but much smaller and produce less dynamic shape changes.
As we outline in the Introduction section at length, cell speed analysis is one important component of studying cell migration. While methods of automated cell tracking exist in commercial software programs, they are not widely used in the field because either they require labeling of cells, or their accuracy and reproducibility (compared to manual tracking) is lacking.47 Many researchers prefer to track unlabeled cells using phase contrast microscopy, both for ease of use and to eliminate added variables. A fully automated system, termed the 2D DIAS, has been developed to study the motility of Dictyostelium amoebae,38,48 but thus far it has been more difficult to develop such a system for epithelial cells, due to their complex behavior and irregular cell shape.39 Ultimately, one of our immediate goals is to update our current manual speed tracking method to include a similar automated system, but not at the expense of accuracy.
Surface area analysis is also an important component of cell migration studies because cell size can be linked to cell shape and health. In general, cells that have suffered mild insults shrink in size as one of the first steps in the apoptotic pathway.49,50 Differentiation of cell lines is also often associated with a change in cell size that may be reflected in our surface area measurements.51,52 There are currently a number of available methods to obtain surface area measurements through image analysis. However, as we previously discussed in the Introduction, many of these methods rely on either the use of fluorescent labeling of cells, differential interference contrast (DIC) microscopy images, and edge detection methods that require heavy computing power, and also often making assumptions about a general cell shape. For some applications, our method of surface area estimation will work well for eukaryotic cells. It is not as accurate as some methods referenced above, but it shows relative changes in surface area very well for phase contrast images, and with very little processing power needed for our algorithm. In addition, our method allows a researcher to follow surface area changes over time (results not shown).
We believe the introduction of the DECCA measurement is our most significant contribution, as this technique captures cell activity in a way that no other applications we are aware of have demonstrated previously. It is important to note that DECCA measures the protrusive activity of cells, whether or not they actually move in a processive manner (i.e., across a substrate). We mention a number of examples previously in this text that would display such behavior (e.g., mutant cells). In fact, a DECCA index need not correlate positively with movement; any positive correlation with cell speed is an indication of how efficient cell activity is towards actual migration. A simple example includes comparison of a motile cell that physically moves across a field, compared to a non-motile cell. The moving cell will always have a DECCA value, since its “footprint” changes from frame to frame. However, the non-moving cell may have a low or high DECCA, depending on the protrusion activity of particular cell. The cell may be completely inactive (if all images are the same, DECCA = 0), or it may change shape without moving its nucleus. by lamellapodial ruffling or creating numerous cell protrusions that lead to shape change. As a result, the image pixel intensity changes, even though their nucleus does not move. A manual version of differential imaging was previously shown in a publication by Fukui et al., and was referred to as producing “difference pictures.”53 However, we are unaware of any other algorithms/computational methods that reflect the same activity as DECCA. Originally, DECCA was developed with the intent to distinguish between two cell types that have the same migration speed, but very different membrane protrusion dynamics (to be included in future work). For example, our analysis demonstrated that although HT-1080 cell speed was significantly altered (p < 0.01) by changing the matrix, the surface area and DECCA of these same cells were essentially unaltered (p > 0.05). This data may indicate that HT-1080 cells have the ability to spread and become activated by both matrices, but for reasons yet to be determined, the cells have a significantly slower migration rate on Fn. Although we cannot explain these differences based on our preliminary analysis, our assay was able to provide additional insight, which would have been missed using population-based cell migration techniques or classical motility tracking assays. By understanding the interplay between cell speed, surface area and DECCA measurements, our method may lead to additional cell migration hypotheses, and thus additional findings.
Knowledge of the fundamental biological mechanisms of cell motility is currently spurring the development of novel pharmacological and genetic approaches that attempt to harness this process, in order to ultimately overcome pathological events such as cancer metastasis. Researchers have screened thousands of compounds for the ability to inhibit cell migration, in hopes of developing new drug targets.54,55 However, commonly used assays that study these interactions cannot distinguish off-target effects. For example, adding formaldehyde to cells would surely halt their cell migration, but will do so by fixing and killing the cells, not because it is a specific inhibitor of migration. In some instances, applying a DECCA measurement may be a useful control for cell health, because of its high resolution and focus on individual cell parameters. However, in order to use our method for large scale screens such as these, some parameters of our experiment will need to be improved upon. We are actively developing many other aspects of this method that can facilitate more efficient data collection and analysis. The most notable addition needed is fully-automated cell tracking software as we mention earlier. We are currently working on a new cell tracking system using our differential imaging method to develop a completely automated technique for epithelial cells similar to those for amoebae.
There are also two known additions needed to track cells using our algorithms: (1) a center of mass calculation on the surface area images, and (2) a center of mass calculation on the DECCA images. These calculations may then be used to predict center of motion and center of velocity, respectively. Accurate edge detection can then be combined with center calculations to identify individual cells and necessary regions of interest (ROIs). This method of auto-selection would also help to narrow the ROI to the minimum window size. With our current program, an operator must manually select the ROI. Future versions of the program will auto-select all ROIs by means of a fast, movie-spanning analysis of the SD of pixel intensity both spatially and temporally. By auto-selecting ROIs, we will decrease both image processing time and non-specific background by applying minimally sized ROIs. Other modifications to our image processing programs are also being adopted, including changes to noise reduction, and image normalization methods.
Another powerful addition to this method is the ability to look at speed, surface area and DECCA at all time points over the course of the movies. For our initial proof-of-principle analysis presented here, all data points were averaged over the course of the movie (e.g., one cell speed measurement per cell per movie). In fact, there were 49 individual measurements each for cell speed, surface area and DECCA. We are keenly aware that our method has thus far untapped data present in these measurements. For example, one could study the stop-and-go pattern of cell locomotion or the change of surface area and DECCA over the course of a movie after exposure to a ligand. We are currently developing statistical methods to analyze our data in this manner.
Here, we present a method that produces a multivariate profile for individual cells based on multiple measurements we have collected: cell speed, surface area and DECCA. In this regard, we can generate three dimensional plots, where each data point represents an individual cell (example in Suppl. Fig. 1). In the future, we plan to use this technique to separate interesting sub-populations within specific cell lines using similar statistical techniques that are used for statistical analysis of cell sorting data.56,57 In this manner, we can further dissect the complex mechanisms of cell migration.
Materials and Methods
Cell culture.
Human fibrosarcoma HT-1080 cells (CCL-121) and human epidermoid carcinoma A431 cells (CRL-1555) were purchased from American Type Culture Collection (Manassas, VA). Both lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Life Technologies, Inc., Rockville, MD) supplemented with 10% fetal bovine serum (FBS; Gemini, Irvine, CA) and 1% glutamine/penicillin/streptomycin antibiotics (Life Technologies), and incubated at 37°C in a humidified, 5% CO2, 95% air atmosphere.
Cell preparation.
The laminin (Ln) isoform Ln-332 (1 µg/mL; purified in-house) or human plasma fibronectin (Fn; 10 µg/mL; Millipore, Billerica, MA) was coated on Nunc™ polystyrene, non-tissue culture treated, 6-well microplate dishes (Cole Parmer, Vernon Hills, IL) for 1 h at room temperature (RT). The dishes were then blocked with 5% milk (Regilait, France) in phosphate buffered saline solution (PBS, Invitrogen) for 1 h at 37°C.
Cell lines were trypsinized (TrypLE Select, Invitrogen, Sunnyvale, CA), neutralized with L-15 media (Invitrogen) supplemented with 10% FBS, washed three times in PBS, and resuspended at a density of 2 × 104 in L-15 media supplemented with 10% FBS in 6-well microplates. Cells were allowed to adhere for 1 h within the heated (37°C) microscope chamber.
Time-lapse microscopy.
Time-lapse microscopy was conducted using a Zeiss Axiovert 200M microscope (Zeiss, Thornwood, NY) equipped with a temperature-controlled chamber and an automated x-y-z stage (0.2 µm repeatability). Microscopy was under the control of MetaMorph software (Molecular Devices, Sunnyvale, CA). At the beginning of each experiment (0 h), six fields were manually selected at random from within each well (in duplicate). Each region was focused manually, and the specific x, y and z coordinates for each was saved using MetaMorph's “Multi-dimensional Acquisition” tool. Phase-contrast images were captured automatically every 5 min for 4 h. Following image capture, all 49 individual images from each particular coordinate were combined using MetaMorph to produce image stacks. A sample of a phase contrast movie for an HT1080 cell can be seen in Supplemental Movie 1.
Cell speed quantification.
Image stacks (with 49 slices; sample slice seen in Fig. 1A) were opened in MetaMorph, and the “Track Points” function was used to manually track cells (Fig. 1B). All cells that remained within the field were tracked by following the center of their nucleus. Cells that collided with other cells and dividing cells were included in our analysis (there was no significant difference between touching, non-touching, and dividing cells' speed, results not shown). Tracking data was exported to a Microsoft Excel spreadsheet for storage. The cell speed parameter was finally calculated by averaging all data collected for each cell first, followed by averaging all cells for that sample population, and final presentation of data includes mean ± standard deviation (SD) of that particular set. After manually tracking cell speed, a unique identifier (based on date, cell line, ECM component, microscopy field and cell number) was assigned to every cell, by manually inserting text next to each cell in the final frame of each movie (see Fig. 1A). This ID system was applied for simplification of binning cells prior to multivariate analysis.
Computer-assisted quantitative analysis (surface area and DECCA).
All subsequent image analysis was performed on non-compressed, 16-bit, TIFF image stacks. Computational and programming support was provided by MathWorks™ MATLAB® (Natick, MA). We used both custom-written algorithms and several advanced MATLAB image-processing toolbox functions, further described below. For each field's stack of microscopic images, a user-defined, rectangular ROI was manually drawn around each cell to be analyzed, and all pixels in each ROI were systematically processed one at a time by the software (Fig. 1C). Subsequently, using two separate, custom-written algorithms for MATLAB, two processed image sequences were produced: (1) intensity-weighted thresholded images (Fig. 1D, used to calculate surface area) and (2) differential intensity images (Fig. 1E). These two image sequences were then used to derive measurements for cellular surface area and DECCA (Fig. 1F), respectively, as described in detail below.
Surface area quantification.
Intensity-weighted, thresholded images were generated using a custom-written MATLAB thresholding algorithm that separates pixels of the background from pixels within the cells based on intensity (Fig. 4A). Temporal SD's were calculated for each pixel, which resulted in an array of SD's for each image stack. A histogram of these values was automatically created and the peaks of each determined by a standard MATLAB “Imaging Toolbox” function. Each peak of the histogram represents the mean for each different pixel class, with the lowest peak representing the background of each image, which was applied as the threshold value for our images. All pixels at the same row and column (i.e., in the same image) that were an SD of intensity lower than the background peak were subsequently saved to a list in Microsoft Excel. When complete, this list represents the intensity characteristic of the background for each image stack. This mean value of each stack (taken from the list) was then subtracted from each pixel in each movie, in order to set the new mean of the background to approximately zero. This leveled image stack was further thresholded and all pixel values within 1.7 SD of the background mean were again reduced to zero. This particular number (1.7) was optimized for our cell types and images, and may vary considerably if the user includes different cell lines or image-capture techniques. In other words, algorithms have been fine-tuned for our model, to remove background noise and appropriately normalize all phase contrast images, prior to further quantitative analysis of surface area.
Figure 4.
Surface Area Algorithms for Image Analysis. (A) Custom-written, MATLAB algorithms were developed to measure surface area of cells (in pixels). Blue text indicates an integrated MATLAB function and black is used for all other text and commands. (B) Sample phase contrast images (top row) and corresponding micrographs of thresholded surface area images (middle row) are presented for a small, medium, and large cell. The calculated number of pixels that corresponds to each cell is also listed for reference (bottom row).
After images were thresholded to remove background using the above technique, all remaining non-zero pixels were taken to represent the selected cell (Fig. 4B); the number of pixels quantitated for each ROI represented the SA measurement for the cell of interest. All 49 frames for each individual cell were then averaged to produce a mean surface area measurement ± SD for that cell over time. This measurement was taken for all adherent, healthy cells of all frames that were in focus and recognized by the software; the occurrence of cells not recognized by the software was negligible (results not shown).
Dynamic expansion/contraction of cell area (DECCA) quantification.
In brief, DECCA is an index number for the total amount of cell motion over time. This motion is not correlated directly with cell migration, as a non-moving cell can ruffle its membrane and produce a DECCA value without physically translocating across a surface or substrate. In this way, the DECCA measurement incorporates both cell motility, and cell shape change. This parameter is calculated as described below.
Differential intensity images (see Fig. 1E) were generated using a custom-written arithmetic algorithm in MATLAB that subtracts the pixel intensity value of each pixel from its counterpart in the same row and column in the next frame (Fig. 5A). These differential intensity images show the relative change in pixel intensity (with color-coded scale) from frame to frame (Fig. 5B), and in this way highlight dynamic cell motion. A non-zero value for a differential pixel indicates an intensity change for that particular pixel from the last frame. Images were thresholded by setting all differential pixels with a value lower than 250 to zero. It is important to note that this value (250) was optimized for our cell types and microscopy technique, and may vary if the user includes different cell lines or image-capture techniques. This analysis results in the creation of an image stack with colored pixels representing the change in pixel intensity from one frame to another. The color and pattern of this differential image stack represents the magnitude and area of cellular motion. These differential image stacks can be viewed as movies to observe the dynamics of cellular motion over time (Suppl. Movie 2). We have also developed an index number to quantify the total amount of cellular motion within these movies. To create the index number, the absolute value of each pixel was taken and the differential values for all pixels in each frame were summed to produce the total absolute differential intensity for each frame. This parameter was averaged across each stack to obtain the DECCA measurement presented for each cell (displayed as intensity units). A sample movie of DECCA images can be seen in Supplemental Movie 3. In this manner, DECCA measures the protrusive activity of cells, whether or not they actually migrate processively. A high DECCA value means that a cell has moved during the movie, possibly due to migration, but it may also be due to the additive effect of membrane ruffling and cell shape change over time. When combined with cell speed data, one can determine if the DECCA number corresponds to cell migration or cell shape change.
Data analysis and statistics.
Data analysis was performed using SPSS, version 16 (SPSS, Inc., Chicago, IL). The Shapiro-Wilks test for normality was applied to all data sets for distribution analysis. The Kolmogorov-Smirnov two-sample non-parametric test was subsequently applied to data to check for significant differences (p < 0.05) across various groups (i.e., by cell line and substrate) for all measurements. For follow-up correlation studies, the unique cell identifying numbers were used to manually combine all three sets of data, so that all parameters for each cell were grouped together in analysis. To analyze relationships between the three measurements, Spearman's R correlation coefficients were calculated for all pairs of variables. All data are presented in terms of mean ± standard deviation (with 95% confidence intervals where indicated).
Acknowledgements
We would like to thank Dr. Alissa Weaver for use of her microscope, also thanks to Dr. Yong Wah Kim, Dr. Kevin Seale, Don Berry and Dr. Nicole Bryce for their technical advice. Support for this work was provided by NIH grants CA47858-17A2 and NCI U54CA113007.
Abbreviations
- DECCA
dynamic expansion/contraction of cell area
- DMEM
dulbecco's modified eagle's media
- ECM
extracellular matrix
- FBS
fetal bovine serum
- Fn
fibronectin
- Ln-332
laminin-332
- PBS
phosphate buffered saline
- ROI
region of interest
Note
Supplementary materials can be found at:
Footnotes
Previously published online as a Cell Adhesion & Migration E-publication: http://www.landesbioscience.com/journals/celladhesion/article/6482
References
- 1.Trinkaus JP. Cells into organs: the forces that shape the embryo. Prentice-Hall: Englewood Cliffs, NJ; 1984. [Google Scholar]
- 2.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 3.Sheetz MP. Cell migration by graded attachment to substrates and contraction. Semin Cell Biol. 1994;5:149–155. doi: 10.1006/scel.1994.1019. [DOI] [PubMed] [Google Scholar]
- 4.Chicurel M. Cell migration research is on the move. Science. 2002;295:606–609. doi: 10.1126/science.295.5555.606. [DOI] [PubMed] [Google Scholar]
- 5.Lauffenburger DA. Models for receptor-mediated cell phenomena: adhesion and migration. Annu Rev Biophys Chem. 1991;20:387–414. doi: 10.1146/annurev.bb.20.060191.002131. [DOI] [PubMed] [Google Scholar]
- 6.Quaranta V. Motility cues in the tumor microenvironment. Differentiation. 2002;70:590–598. doi: 10.1046/j.1432-0436.2002.700912.x. [DOI] [PubMed] [Google Scholar]
- 7.Dormann D, Weijer CJ. Imaging of cell migration. EMBO J. 2006;25:3480–3493. doi: 10.1038/sj.emboj.7601227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motilitly by enhancing lamellipodial persistence. Curr Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 9.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JL, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 11.Kellermayer MSZ, Karsai A, Benke M, Soos K, Penke B. Stepwise dynamics of epitaxially growing single amyloid fibrils. PNAS. 2008;105:141–144. doi: 10.1073/pnas.0704305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller-Rath R, Gavenis K, Gravius S, Andereya S, Mumme T, Schneider U. In vivo cultivation of human articular chondrocytes in a nude mouse-based contained defect organ culture model. Bio-Med Mat Eng. 2007;17:357–366. [PubMed] [Google Scholar]
- 13.Jokhadar SZ, Znidarcic T, Svetina S, Batista U. The effect of substrate and adsorbed proteins on adhesion, growth, and shape of CaCo-2 cells. Cell Biol Int. 2007:1–12. doi: 10.1016/j.cellbi.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 14.Friel JJ. Practical guide to image analysis. ASM International. 2000 [Google Scholar]
- 15.Alexopoulos LG, Erickson GR, Guilak F. A method for quantifying cell size from differential interference contrast images: validation and application to osmotically stressed chondrocytes. J Microsc. 2002;205:125–135. doi: 10.1046/j.0022-2720.2001.00976.x. [DOI] [PubMed] [Google Scholar]
- 16.Toraason M, Krueger JA, Busch KA, Shaw PB. Automated surface area measurement of cultured cardiac myocytes. Cytotechnology. 1990;4:155–161. doi: 10.1007/BF00365096. [DOI] [PubMed] [Google Scholar]
- 17.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:1–10. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu-van Opstal WY, Billardon C, Caillaud T, Carvajal-Gonzalez S, Colliot G, Bisconte JC, Rostene W. Automatic cell culture quantitation with TRAKCELL: application to cell toxicology and differentiation. Cell Biol Toxicol. 1994;10:387–392. doi: 10.1007/BF00755787. [DOI] [PubMed] [Google Scholar]
- 19.Ray N, Acton ST. Data acceptance for automated leukocyte tracking through segmentation of spatiotemporal images. IEEE Trans Biomed Eng. 2005;52:1702–1712. doi: 10.1109/TBME.2005.855718. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee DP, Ray N, Acton ST. IEEE Trans Biomed Image Process. 2004;13:562–572. doi: 10.1109/tip.2003.819858. [DOI] [PubMed] [Google Scholar]
- 21.Truskey GA, Proulx TL. Quantitation of cell area on glass and fibronectin-coated surfaces by digital image analysis. Biotechnol Prog. 1990;6:513–519. doi: 10.1021/bp00006a016. [DOI] [PubMed] [Google Scholar]
- 22.Ionescu-Zanetti C, Wang LP, Di Carlo D, Hung P, Di Blas A, Hughey R, Lee LP. Alkaline hemolysis fragility is dependent on cell shape: results from a morphology tracker. Cytometry A. 2005;65:116–123. doi: 10.1002/cyto.a.20135. [DOI] [PubMed] [Google Scholar]
- 23.Roy P, Rajfur Z, Pormorski P, Jacobson K. Microscope-based techniques to study cell adhesion and migration. Nat Cell Biol. 2002;4:91–96. doi: 10.1038/ncb0402-e91. [DOI] [PubMed] [Google Scholar]
- 24.Stephens DJ, Allan VJ. Light microscopy techniques for live cell imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- 25.Guan JL. Cell migration: developmental methods and protocols. Humana Press; 2004. [Google Scholar]
- 26.Bahnson A, Athanassiou C, Koebler D, Qian L, Shun T, Shields D, Yu H, Wang H, Goff J, Cheng T, Houck R, Cowsert L. Automated measurement of cell motility and proliferation. BMC Cell Biol. 2005;6:19. doi: 10.1186/1471-2121-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ariano P, Distasi C, Gilardino A, Zamburlin P, Ferraro M. A simple method to study cellular migration. J Neurosci Meth. 2004;141:271–276. doi: 10.1016/j.jneumeth.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Weaver AM. Invadopodia: specialized structures for cancer invasion. Clin Exp Metastasis. 2006;23:97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- 29.Webb DJ, Brown CM, Horwitz AF. Illuminating adhesion complexes in migrating cells: moving toward a bright future. Curr Opin Cell Biol. 2003;15:614–620. doi: 10.1016/s0955-0674(03)00105-4. [DOI] [PubMed] [Google Scholar]
- 30.DiMilla PA, Quinn J, Albelda SM, Lauffenburger DA. Measurement of individual cell migration parameters for human tissue cells. Aiche J. 1992;38:1092–1104. [Google Scholar]
- 31.Zygourakis K. Quantification and regulation of cell migration. Tissue Eng. 1996:2. doi: 10.1089/ten.1996.2.1. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe S, Hirose M, Wang XE, Maehiro K, Murai T, Kobayashi O, Mikami H, Otaka K, Miyazaki A, Sato N. A new model to study repair of gastric mucosa using primary cultured rabbit gastric epithelial cells. J Clin Gastroenterol. 1995;21:40–44. [PubMed] [Google Scholar]
- 33.Lovelady DC, Richmond TC, Maggi AN, Lo CM, Rabson DA. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:041908. doi: 10.1103/PhysRevE.76.041908. [DOI] [PubMed] [Google Scholar]
- 34.Keese CR, Wegener J, Walker SR, Giaever I. Electrical wound-healing assay for cells in vitro. PNAS. 2004;101:1554–1559. doi: 10.1073/pnas.0307588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingber DE. Fibronectin controls capillary endothelial growth by modulating cell shape. Proc Natl Acad Sci USA. 1990;87:3579–3583. doi: 10.1073/pnas.87.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang N, Ingber DE. Control of cytoskeletal mechanics by extracellular matrix, cell shape, and mechanical tension. Biophys J. 1994;66:2181–2189. doi: 10.1016/S0006-3495(94)81014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soll DR. The use of computers in understanding how animal cells crawl. Int Rev Cytol. 1995;163:43–104. [PubMed] [Google Scholar]
- 38.Wessels D, Kuhl S, Soll DR. Application of 2D and 3D DIAS to motion analysis of live cells in transmission and confocal microscopy imaging. Methods Mol Biol. 2006;346:261–279. doi: 10.1385/1-59745-144-4:261. [DOI] [PubMed] [Google Scholar]
- 39.Zimmer C, Olivo-Marin JC. Coupled parametric active contours. IEEE Trans Pattern Anal Mach Intell. 2005;27:1838–1842. doi: 10.1109/TPAMI.2005.214. [DOI] [PubMed] [Google Scholar]
- 40.Soll DR, Voss E, Varnum-Finney B, Wessels D. “Dynamic Morphology System”: a method for quantitating changes in shape, pseudopod formation, and motion in normal and mutant amoebae of Dictyostelium discoideum. J Cell Biochem. 1988;37:177–192. doi: 10.1002/jcb.240370205. [DOI] [PubMed] [Google Scholar]
- 41.Zimmer C, Bo Zhang, Dufour A, Thebaud A, Berlemont S, Meas-Yedid V, Marin JCO. On the digital trail of mobile cells. Signal Processing Magazine, IEEE. 2006;23:54–62. [Google Scholar]
- 42.Hinz B, Alt W, Johnen C, Herzog V, Kaiser H. Quantifying lamella dynamics of cultured cells by SACED, a new computer-assisted motion analysis. Exp Cell Res. 1999;251:234–243. doi: 10.1006/excr.1999.4541. [DOI] [PubMed] [Google Scholar]
- 43.Cohen J. Statistical power analysis for the behavioral sciences. 2nd ed. Hillsdale, NJ: Lawrence Erlbaum Associates; 1988. [Google Scholar]
- 44.Winterwood NE, Varzavand A, Meland MN, Ashman LK, Stiff CS. A critical role for tetraspanin CD151 in alpha3beta1 and alpha6beta4 integrin-dependent tumor cell functions on laminin-5. Mol Biol Cell. 2006;17:2707–2721. doi: 10.1091/mbc.E05-11-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng H, Briske-Anderson M, Idso JP, Hunt CD. The selenium metabolite methylselenol inhibits the migration and invasion potential of HT1080 cells. J Nutr. 2006;136:1528–1532. doi: 10.1093/jn/136.6.1528. [DOI] [PubMed] [Google Scholar]
- 46.Dove A. Membrane Specialization. J Cell Biol. 1999;145:2. [Google Scholar]
- 47.Chon JH, Vizena AD, Rock BM, Chaikof EL. Characterization of single-cell migration using a computer-aided fluorescence time-lapse videomicroscopy system. Anal Biochem. 1997;252:246–254. doi: 10.1006/abio.1997.2321. [DOI] [PubMed] [Google Scholar]
- 48.Wessels JT, Hoffman RM, Wouters FS. The use of transgenic fluorescent mouse strains, fluorescent protein coding vectors, and innovative imaging techniques in the life sciences. Cytometry A. 2008;73:490–491. doi: 10.1002/cyto.a.20548. [DOI] [PubMed] [Google Scholar]
- 49.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aharon R, Bar-Shavit Z. Involvement of Aquaporin 9 in Osteoclast Differentiation. J Biol Chem. 2006;281:19305–19309. doi: 10.1074/jbc.M601728200. [DOI] [PubMed] [Google Scholar]
- 52.Zouboulis CHC, Krieter A, Gollnick H, Mischke D, Orfano CE. Progressive differentiation of human sebocytes in vitro is characterized by increasing cell size and altering antigen expression and is regulated by culture duration and retinoids. Exp Dermatol. 1994;3:151–160. doi: 10.1111/j.1600-0625.1994.tb00271.x. [DOI] [PubMed] [Google Scholar]
- 53.Fukui Y, Murray J, Riddelle KS, Soll DR. Cell behavior and actomyosin organization in Dictyostelium during substrate exploration. Cell Struct Funct. 1991;16:289–301. doi: 10.1247/csf.16.289. [DOI] [PubMed] [Google Scholar]
- 54.McHenry KC. A non-antibacterial oxazolidinone derivative that inhibits epithelial cell sheet migration. Chembiochemistry. 2002;3:1105–1111. doi: 10.1002/1439-7633(20021104)3:11<1105::AID-CBIC1105>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 55.Yarrow JC, Totsukawa G, Charras GT, Mitchison TJ. Screening for cell migration inhibitors using automated microscopy reveals a new rho-kinase inhibitor. Chem Biol. 2005;12:385–395. doi: 10.1016/j.chembiol.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 56.Bindschadler M, McGrath JL. Sheet migration by wounded monolayers as an emergent property of single cell dynamics. J Cell Sci. 2007;120:876–884. doi: 10.1242/jcs.03395. [DOI] [PubMed] [Google Scholar]
- 57.Mochizuki A, Iwasa Y, Takeda Y. A Stochastic model for cell sorting and measuring cell-cell adhesion. J Theo Biol. 1996;179:129–146. [Google Scholar]