Abstract
Papillary thyroid carcinoma (PTC) displays higher heritability than most other cancers. To search for genes predisposing to PTC, we performed a genome-wide linkage analysis in a large family with PTC and melanoma. Among several peaks the highest was at 8q24, with a maximum nonparametric linkage (NPL) score of 7.03. Linkage analysis was then broadened to comprise 25 additional PTC families which produced a maximum NPL score of 3.2, p = 0.007 at the 8q24 locus. Fine mapping with microsatellite markers was compatible with linkage to the 8q24 locus in 10 of the 26 families. In the large family a ~320 Kb haplotype was shared by individuals with PTC, melanoma, or benign thyroid disease, but not by unaffected individuals. A 12 Kb haplotype of 8 SNP markers within the larger haplotype was shared by 9 out of the 10 families in which the 8q24 locus was compatible with linkage. The shared haplotype is located within 2 known overlapping protein-coding genes, thyroglobulin (TG) and Src-like adaptor (SLA). Resequencing of the coding and control regions of TG and SLA did not disclose putative mutations in PTC patients. Embedded in the TG-SLA region are three likely non-coding RNA genes, one of which (AK023948) harbors the 8-SNP haplotype. Resequencing of AK023948 and one of the other RNA genes did not reveal candidate mutations. Gene expression analysis indicated that AK023948 is significantly downregulated in most PTC tumors. The putative non-coding RNA gene AK023948 is a candidate suseptibility gene for PTC.
Keywords: papillary thyroid carcinoma, susceptibility locus, haplotype sharing, non-coding RNA gene
Introduction
It is a relatively little discussed fact that non-medullary thyroid carcinoma (NMTC) shows a high degree of heritability. Several large case control studies have reported the heritability of NMTC to be one of the highest of all cancers. Papillary thyroid carcinoma (PTC) is the main form of NMTC, accounting for approximately 80% of all thyroid cancers. PTC is mostly sporadic; however increasingly over the past 20 years, the occurrence of PTC running in families has been observed (1). It has been estimated that 5 to 10% of all PTC are “familial” (2, 3). To study the degree of heritability/familiality of cancers, large case control studies have been published from Utah and Sweden (3, 4). An overall familial risk ratio (FRR) of approximately 2 for all cancers combined suggests a clear but modest overall contribution of predisposing genes to cancer (5). In the most common cancers, in which well-characterized subsets are known to be inherited as regular Mendelian traits (colorectal, breast), the FRRs were mostly between 2 and 4. In contrast, the highest FRR was noted for thyroid cancer (3, 4) (FRR 8.48 in Utah, 9.51 in Sweden). Other case control studies have measured relative risk in first degree family members of probands with thyroid cancer specifically excluding the medullary form obtaining similar results in that PTC was among the cancers with strongest heritability (4, 6, 7). In conclusion, a major hereditary component is apparent in the predisposition to PTC.
Several linkage analyses in PTC families have been published in the past reporting at least four genomic regions (on chromosomes 1q21, 2q21, 14q31, and 19p32) which may harbor predisposing genes (8–11). However, no such predisposing genes have been identified.
In this study, we conducted linkage analyses with high density SNP arrays in PTC families to identify candidate regions. We initially studied a large three-generation family with PTC and melanoma, then enlarged the study to comprise 26 PTC families. Evidence from linkage, haplotype sharing, and gene expression analysis implicated a putative non-coding RNA gene (AK023948 in 8q24) as a candidate gene for PTC predisposition.
Materials and Methods
The studies were approved by the Institutional Review Board at the Ohio State University, and all subjects gave written informed consent before participation.
Family samples and genomic DNA extraction
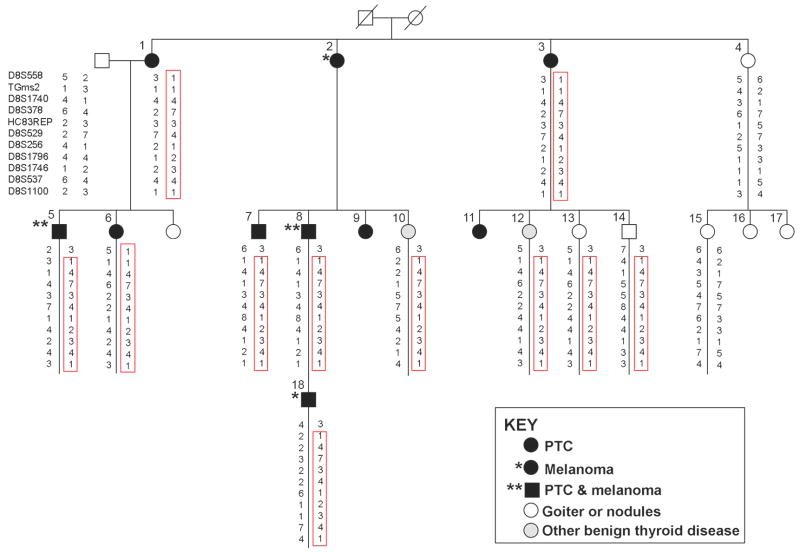
The key family in this study shown in Fig. 1. comprised individuals affected with PTC and melanoma (family #1). There were 8 individuals affected with PTC; two of them had both PTC and melanoma. Among the remaining family members, 2 had melanoma only and 2 had chronic lymphocytic leukemia. An additional 10 individuals had benign thyroid disease (nodules or goiter), including one individual with goiter who also had both cutaneous and ocular melanoma, as well as breast cancer. An additional 25 families with at least 2 confirmed cases of non-medullary thyroid cancer in close relatives were recruited. The majority (22 of 25) had 3 or more affected individuals, including a large family with 13 members affected with PTC (family #21). Family history information, pathology reports confirming the diagnosis of thyroid cancer or thyroid disease, as well as blood and tissue samples were collected from all consenting affected individuals and key unaffected individuals. The pedigrees of the 25 kindreds are provided in Supplementary Fig. 1. Genomic DNA was extracted from blood according to standard phenol-chloroform extraction procedures.
Figure 1.
Haplotypes of microsatellite markers in members of family #1. A unique haplotype (boxed) co-segregates with PTC, melanoma, and some benign thyroid diseases.
Genotyping
Genome-wide analysis of single nucleotide polymorphisms (SNPs) was performed by using the Affymetrix GeneChip Human Mapping 50K Array (50K_Xba_240 chip), or Affymetrix GeneChip Human Mapping 500K (Nsp 250K and Sty 250K) arrays. Sample preparation, chip hybridization and data quality controls were carried out according to Affymetrix’ recommendations. SNP genotype calls were made with Genechip Genotyping Analysis Software (GTYPE) 4.0 (Affymetrix) with default parameters or using theBRLMM program from Affymetrix. The SNP call rate was over 92% with a p value of 0.3. The Mendelian error rate was below 0.2% and errors were removed before analysis.
Genotyping with microsatellite markers
Microsatellite markers were picked to span the linkage peak region on 8q24 based on the NCBI-uniSTS-deCode database1 or markers described in the literature. The PCR primers flanking the microsatellites were obtained from the NCBI-uniSTS database or designed with the Primer3 program. Microsatellite marker designations and the PCR primer sequences are provided in Supplementary Table 1. The PCR assays were performed according to the standard PCR protocol except that one PCR primer was labeled with a fluorescent dye (HEX, FAM, or TET). Most frequently the PCR assays were carried out using the following conditions: 2 min at 94 C; followed by 30 cycles of 30 s at 94 C, 30 s at 58 C, and 30 s at 72 C; followed by a final extension of 10 min at 72 C. The allele analysis was performed by using ABI 3730 DNA Analyzer.
Statistical analysis
For genome-wide nonparametric linkage analysis, MERLIN (12) was employed. Calculated allele frequencies based on genotyped individuals were used for NPL scoring. Genetic positions of NPL scores on a chromosome were indicated by using the deCODE map retrieved from Affymetrix NetAffx. The data set from family #1 was also analyzed with GENEHUNTER 2.1 (13) software with randomly selected SNPs using both non-parametric and parametric methods. Allele frequencies were calculated based on all genotyped individuals in the dataset. The haplotypes were constructed by using GENEHUNTER 2.1. or MERLIN, and Haplopainter. The shared haplotype for family #1 and 9 other families was constructed based on markers in the linkage peak region. Haplotype construction and frequency estimation in healthy controls was performed with PHASE V2.1.1 program (14, 15).
Thyroid samples and genomic DNA and RNA extraction
Thyroid paraffin blocks were collected from 3 PTC individuals (#5, #6, #9) in family #1. Fresh frozen thyroid samples were collected from one individual in each of 2 families (individual #1 in family #3 and individual #2 in family #8) (Supplementary Table 2). Fresh snap-frozen thyroid tissue was obtained from patients with sporadic PTC undergoing surgical resection, including tumor tissue (T-PTC, n=26) and normal thyroid tissue from the same gland (N-PTC, n=26). Control “normal” thyroid tissue (N-Thy, n=4) was collected from consenting individuals who had surgery because of laryngeal malignancy but no thyroid disease. Clinical data and information on the specimens will be provided upon request. These freash frozen tissues were collected and selected for study after histological examination. Genomic DNA was extracted by a standard phenol-chloroform procedure. Total RNA from fresh frozen tissue was extracted with TRIzol Reagent (Invitrogen). Total RNA from paraffin blocks was extracted using the RecoverAll Total Nucleic Acid Isolation kit (Ambion).
Semi-quantitative and quantitative RT-PCR
Total RNA was first treated with DNase-1 (Ambion) and then reverse transcribed to cDNA with the SuperScript First Strand Synthesis system (Invitrogen). Candidate genes and an endogenous control gene, glyceraldeyde-3-phosphate dehydrogenase (GAPDH) were included in the same PCR reaction for semi-quantitative RT-PCR. All PCR assays were verified to be in the linear range by testing with different cycle numbers. Quantitative real-time PCR was performed by using an ABI PRISM 7700 DNA Sequence Detection System (Applied Biosystems) and a SYBR Green PCR kit (Applied Biosystems). The comparative threshold cycle method was used to calculate the relative gene expression. Primers for amplification of AK023948 and GAPDH are listed in Supplementary Table 1.
DNA Sequencing
Genomic DNA from one PTC patient in each selected family (2 PTC patients from family #1) was used for resequencing. Dideoxy-DNA sequencing of candidate genes was performed after PCR amplification of genomic DNA. The PCR primers and PCR conditions are available upon request. PCR products were directly sequenced with one of the PCR primers or primers specifically designed. Automated sequencing was performed using a PE373 DNA sequencer.
Results
Genome-wide linkage analysis in a large PTC and melanoma family
The initial genome-wide linkage analysis was conducted in a single large PTC and melanoma family (#1) using the Affymetrix 50K SNP array. Blood DNA samples from 15 individuals were genotyped, including 7 affected (PTC, and/or melanoma) individuals. Separate linkage analyses were carried out using two phenotypic designations, one assigning affected status to only the 7 individuals with PTC and/or melanoma, and one in which these 7 individuals plus 4 additional family members with benign thyroid disease were scored as affected. At least 4 prominent linkage peaks were identified. The most highly ranked peak was located on chromosome 8, from 142.70 to 154.53 cM (deCODE) (8q24 locus), with a maximum NPL score of 7.03 in cancer cases only and 7.1 if benign thyroid disease cases were included. The peaks from the two analyses overlapped fully (data not shown). In addition, 3 other chromosomal loci, 6q27, 12q21 and 14q11, displayed NPL scores from 6.5 to 6.9. The details of these 4 linkage peaks are described in Table 1.
Table 1.
Putative susceptibility loci in a large PTC and melanoma family
| Chromosome | Max NPL* | SNP markers | Position (bp) | size (Mb) |
|---|---|---|---|---|
| 8q24 | 7.03 | SNP_A-1705523 | 133839469 | 6.3 |
| SNP_A-1730589 | 140093351 | |||
| 12q21-24 | 6.94 | SNP_A-1696322 | 77766365 | 30.3 |
| SNP_A-1667888 | 108153034 | |||
| 14q11 | 6.75 | SNP_A-1720255 | 19285288 | 4.2 |
| SNP_A-1720255 | 23471589 | |||
| 6q27 | 6.5 | SNP_A-1702193 | 164689010 | 3.4 |
| SNP_A-1727250 | 168112476 |
Max NPL: maximum non-parametric linkage score obtained by linakge analysis with Affymetrix GeneChip Human Mapping 50K Array.
We noticed that the thyroglobulin (TG) gene is located in the 8q24 region of linkage, and the 8q24 locus overlaps with a reported susceptibility locus in autoimmune thyroid diseases (16). These findings prompted us to examine the TG region in 8q24 more closely.
To refine the 8q24 locus, we genotyped 11 microsatellite markers (D8S558, TGms2, D8S1740, D8S378, HC83REP, D8S529, D8S256, D8S1796, D8S1746, D8S537, D8S1100) in family #1. A haplotype was identified in 7 individuals with PTC alone, PTC and melanoma, or melanoma alone, and 4 individuals with benign thyroid disease (Fig. 1). One individual (#4) with thyroid nodules did not have this haplotype. For further linkage analysis, the microsatellite markers were combined with SNP markers from the Affymetrix 50K array. Nonparametric multipoint linkage analysis with the combined markers produced a NPL score of 7.0, a LOD score of 1.9, and a linkage interval of approximately 6.2 Mb (Supplementary Fig. 2).
The 8q24 locus in 26 PTC families
Genome-wide linkage analysis was widened by testing an additional 25 PTC families using the Affymetrix GeneChip Human Mapping Nsp250K array. We generated about 250,000 SNP genotypes for 135 individuals, including 86 individuals affected with PTC and 13 obligate carriers.
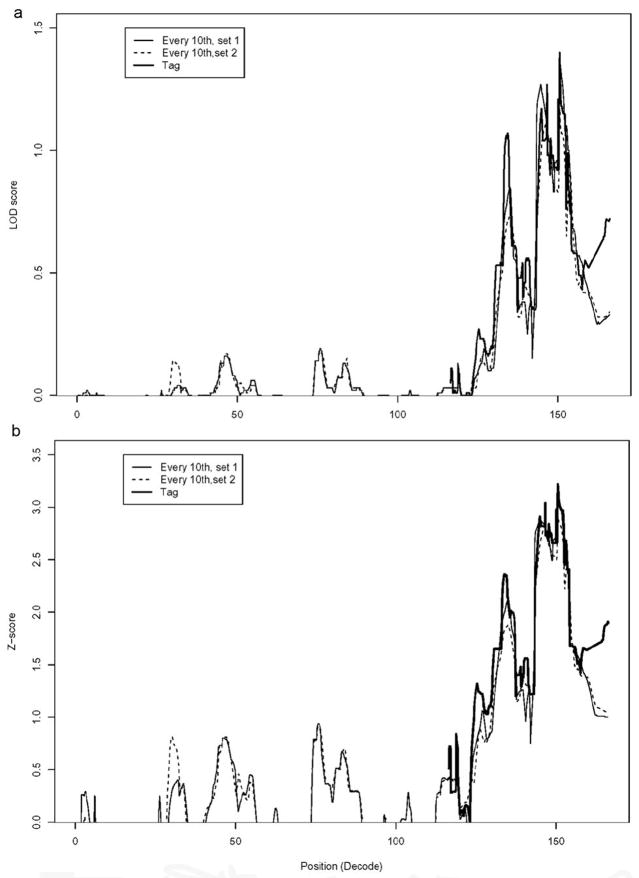
To deal with the large amount of genotyping data and to minimize spurious co-segregation due to linkage disequilibrium between adjacent markers, the genome wide multipoint non-parametric linkage analysis was performed with MERLIN software with different sets of selected SNP markers. We performed three independent parallel analyses using the following SNP sets: 1 out of every 10 or 12 SNPs; SNPs with the highest heterozygosity frequency and a distance of +/− 500Kb between each consecutive marker; and a randomly selected set of SNPs. Genome-wide linkage analysis in all 26 families produced several regions positive for linkage, including the 8q24 locus. The overall linkage results in these families will be summarized and published elsewhere.
To compare the linkage results using different sets of SNP markers, we plotted NPL scores and LOD scores for chromosome 8. As can be seen in Fig. 2 the overlaid plots are similar. The combined linkage analysis in the 26 families produced a maximum NPL score of 3.2, p value 0.007, and a maximum of LOD score of 1.3, p value 0.02. These families could be divided into three groups: families with NPL score >= 1 (n=9), which are compatible with linkage to the 8q24 locus; families with NPL score =<0 (n=10), which are likely not linked to this locus; and families with intermediate NPL scores between 0.2 to 0.7 (n=7). One of these (family #20) with an NPL score of 0.7 also produced positive evidence of linkage to the 8q24 locus by microsatellite marker genotyping (data not shown), and was included in the group of families being compatible with 8q24 linkage (n=10), and further studied. The clinical information of these 10 families is summarized in Supplementary Table 2. Among the 10 families, 3 additional families (excluding family #1) had individuals with melanoma (ranging from 1–3 cases of cutaneous melanoma/family, as well as one case of uveal melanoma). None of these individuals had both PTC and melanoma. We noticed there was at least one individual with autoimmune thyroid disease (AITD) in 9 out of the 10 families, including 6 cases in family #1 (Supplementary Table 2). Of the 18 individuals with AITD in these 10 families, 10 had a subsequent or concurrent diagnosis of PTC.
Figure 2.
NPL and LOD score plots of chromosome 8 from 26 PTC families. Multi-point nonparametric linkage analysis is shown with two different sets of SNPs separately. The linkage peak region was further re-analyzed with tag SNPs only; the tag SNPs (r=1.0) were chosen by using tagger program (http://www.broad.mit.edu/mpg/tagger/). The LOD score plots (a) and NPL score plots (b) with 3 sets of SNP markers are shown.
Haplotypes
Haplotypes for the 8q24 locus were constructed for family #1 based on markers in the central peak region. A 10-marker haplotype consisting of 6 SNPs from the 50K chip and 4 microsatellite markers was shared by 7 individuals affected with PTC and/or melanoma and 4 individuals with benign thyroid disease, but not by any of the unaffected individuals who we genotyped. The haplotype was reconstructed with combined SNPs from the Affymetrix 50K chip and 250K Nsp chip and refined to a region of ~ 320 kb in the TG gene region (data not shown). Further haplotype sharing analysis across the 10 families showing positive linkage to 8q24 identified a smaller ~12 kb haplotype within the larger haplotype; this small haplotype was composed of 8 SNPs; the genotypes of these SNPs were obtained either from the Affymetrix 500K array, or by direct sequencing of the region. This small haplotype was shared by at least 9 families in which the 8q24 locus was compatible with linkage (Table 2). This haplotype was seen in PTC individuals from other PTC families as well as in healthy controls. The frequency of the haplotype was 40% in our 74 Caucasian control samples.
Table 2.
A shared haplotype among PTC families
| PTC family ID** |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dbSNP | SNP ID in chip* | Position (bp) | Gene | 1 | 6 | 20 | 3 | 12 | 10 | 24 | 9 | 8 | 17 |
| rs4736618 | SNP_A-2036048 | 134129641 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | |
| rs2252696 | SNP_A-1995222 | 134132714 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| rs1533910 | SNP_A-4213261 | 134133334 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | |
| rs9942754 | SNP_A-1821028 | 134133815 | C | C | C | C | C | C | C | C | C | C | |
| rs2702969 | SNP_A-4300382 | 134136714 | AK023948 | C | C | C | C | C | C | C | C | C | C |
| rs2702968 | n/I*** | 134137551 | AK023948 | A | A | A | A | A | A | A | A | A | A |
| rs2702967 | n/I | 134137930 | AK023948 | A | A | A | A | A | A | A | A | A | G |
| rs2739136 | n/I | 134138374 | AK023948 | A | A | A | A | A | A | A | A | A | A |
| rs17695552 | n/I | 134138602 | AK023948 | T | T | T | T | T | T | T | T | T | T |
| rs2702966 | SNP_A-4291953 | 134139958 | C | C | C | C | C | C | C | C | C | C | |
| rs2741205 | SNP_A-1789153 | 134145907 | G | G | G | G | G | G | G | G | G | A | |
| rs2248346 | SNP_A-2244627 | 134145946 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| rs872643 | SNP_A-4231278 | 134160784 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | |
| rs2739169 | SNP_A-4216065 | 134165098 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | |
The SNP ID in Affymetrix Nsp and Sty 500K chip.
Family IDs of 11 PTC families which showed linkage to the 8q24 locus
The co-segreagting allele in each family is shown; the shared haplotype from 8 SNPs is indicated by bolded letters.
n/I: not in the chips.
Genes in the region
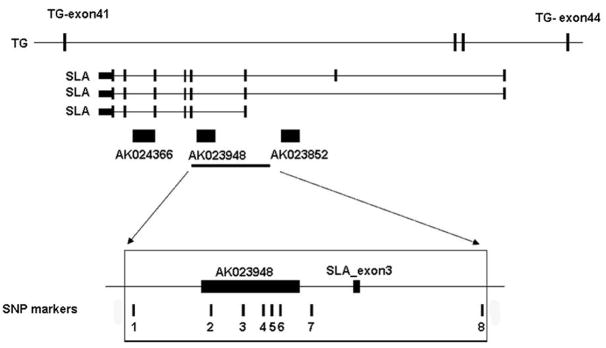
The shared haplotype is located within 2 known coding genes, TG and Src-like adaptor (SLA), and also covers one uncharacterized cDNA, AK023948. SLA is encoded by the antisense strand of three introns of the TG gene. The cDNA AK023948 and two other cDNAs (AK023852 and AK024366) reside in the introns of the SLA gene; These cDNAs were annotated as single-exon transcripts of 2.1 kb to 2.8 kb, which are likely non-coding RNA genes based on the very small predicted open reading frame (less than 100 amino acids in each), with no known paralogs or orthologs in other species. A diagram of the genomic structure of this region is shown in Fig. 3. It is noteworthy that the two cDNAs (AK023948 and AK023852) and several ESTs aligned with these cDNAs were isolated from thyroid tissue. The 8 SNP markers in the haplotype are indicated in Fig. 3.
Figure 3.
The thyroglobulin (TG) gene cluster and shared haplotype in the 8q24 locus. The cluster consists of the large TG gene (48 exons total); the SLA gene encoded by the opposite strand in TG introns 41, 42 and 43; and three cDNAs encoded in introns of SLA. The relative positions of the SNP markers forming a shared haplotype in 9 families was based on UCSC Genome Browser (http://genome.ucsc.edu/). The 8 SNPs forming the shared haplotype are: 1: rs9942754; 2: rs2702969; 3: rs2702968; 4: rs2702967; 5: rs2739136; 6: rs17695552; 7: rs2702966; 8: rs2741205.
Sequencing TG, SLA, AK023948, and AK023852
Using genomic DNA from blood we resequenced the TG, SLA, AK023948, and AK023852 genes in 7 PTC patients from 6 selected families; one patient from each family but two patients from family #1 (individuals #1 and #8 in Fig. 1). All exons and exon-intron boundary regions of the 4 genes were sequenced as well as the 2 kb promoter region of SLA isoform 1 (NM_006748) and the 2 additional exons of the 2 other SLA isoforms (NM_001045556 and NM_001045557). Numerous DNA polymorphisms were found (data not shown) but no changes suggestive of deleterious mutations were seen.
Expression analysis of genes within the locus
We have previously established gene expression profiles in PTC by using Affymetrix HG-U133 plus 2 array ((17, 18). Our array data indicated that the expression level of AK023948 and AK023852 was 5 fold lower in PTC tumor vs. normal thyroid tissue in approximately half of the samples tested, while notable differences in gene expression levels of the TG and SLA genes were not obvious between tumor and unaffected tissue (Supplementary Table 3).
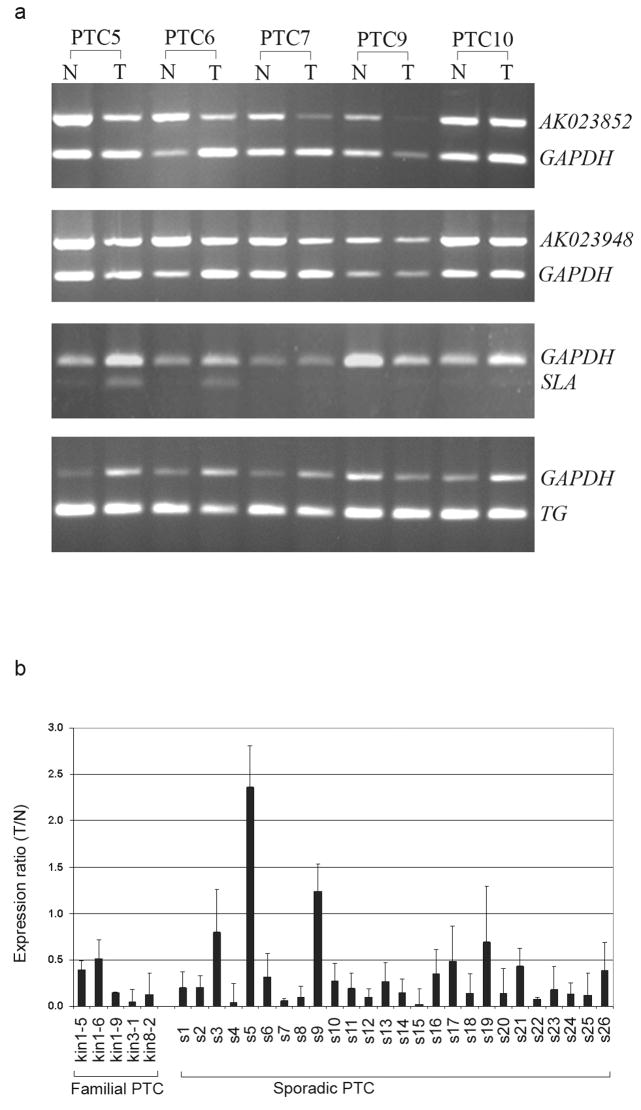
To verify the array data we performed semi-quantitative RT-PCR and confirmed that AK023948 and AK023852 were significantly downregulated in some sporadic PTC tumors as seen in samples PTC6 and PTC7 (Fig. 4a). The expression patterns of TG and AK024366 were assayed in the same sample set, with no notable alteration in tumor vs. normal (Fig.4a and data not shown). Expression of the SLA gene was very low and hard to detect in most of the samples tested but in at least 2 cases (PTC5 and PTC6) expression was slightly higher in the tumor than in unaffected tissue (Fig. 4a). Quantitative real time RT-PCR data (Figure 4b) showed that the expression level of AK023948 was low in 5 cases of familial PTC. There were 3 cases from the large family (kin1); samples kin1-5 and kin1-6 showed about 50% decrease (T/N ratios 0.42, 0.51, respectively) while sample kin1-9 showed a remarkable decrease (T/N ratio 0.14). Two other familial samples (kin3-1 and kin8-2) showed significant downexpression (0.05 and 0.13, respectively). We also tested 26 cases of sporadic PTC with real time RT-PCR (Fig. 4b). Most of the tumors (24 out of 26) showed downexpression (T/N ratios from 0.02 to 0.8), and two samples showed over-expression (T/N ratios: 1.23 and 2.36) (Fig.4b). Overall, the downexpression of the AK023948 gene was strong.
Figure 4.
Gene expression analysis in PTC. (a) Semi-quantitative RT-PCR of AK023852, AK023948, SLA, and TG in 5 sporadic PTC samples from which tumor (T) and paired unaffected thyroid tissue (N) were analyzed. GAPDH was used as an internal control. (b) Quantitative real time RT-PCR of AK023948 in familial and sporadic PTC samples. GAPDH was used as control. The expression ratio (T/N) of PTC tumor (T) versus paired unaffected normal (N) was calculated by the comparative threshold cycle method (mean and the standard deviation are shown).
Discussion
Linkage analysis is the typical first step to localize disease genes. In PTC at least 4 susceptibility loci have been proposed by others based on linkage; however no genes have been identified. A locus on 19p32 was identified in a French family with multiple cases of an unusual form of thyroid tumors with cell oxyphilia (8). In a large Canadian family with 18 cases of multinodular goiter and 2 cases of PTC a locus on 14q31 was proposed (9). A 1q21 locus was identified in one family with PTC and renal papillary cancer (10). Finally, a 2q21 locus was disclosed through a large study including 191 members of 80 families (11). The proportion of all families showing linkage to that locus was estimated at as high as 36%. Intriguingly, by analyzing 10 families (9 with the oxyphilic subtype), including the large French family in which the 19p32 locus was originally identified, evidence of linkage was seen not only to the 19p32 locus, but to the 2q21 locus as well (19). We surmise that these previous findings suggest a high degree of genetic heterogeneity and possible multigenic inheritance in PTC.
We recruited a large three-generation family with individuals affected with PTC and melanoma and other types of cancer. Our initial genome-wide linkage analysis considering individuals with PTC and melanoma as “affected” disclosed four candidate genomic regions, on chromosomes 8q24, 6q27, 12q21-24, and 14q11. The existence of multiple linkage peaks found in one single family is hard to explain as regular genetic heterogeneity and it is possible that some of the peaks represent random noise. Epidemiological studies have provided evidence that there is an elevated thyroid cancer risk among cutaneous melanoma survivors (20–22). PTC and melanoma may have overlapping genetic pathways in that a high prevalence of BRAF mutation has been seen in both types of tumors (23–25). It is therefore possible that undisclosed pathogenetic pathways, even susceptibility gene(s), may be shared between PTC and melanoma (25). Characteristics of familial PTC include an association with benign nodular thyroid disease. Interestingly, the linkage at the 8q24 locus in the large family was not only found in individuals affected with PTC and melanoma, but also in four individuals affected with benign thyroid disease. This observation suggests that the 8q24 locus could harbor one or several susceptibility genes for PTC and other benign thyroid disease or that within this particular family, the two conditions are related. Combined linkage analysis in 26 PTC families and additional fine mapping with microsatellite markers provided evidence compatible with linkage to the 8q24 locus in 10 PTC families, suggesting that the 8q24 locus may contribute to a sizeable portion, but not the majority, of cases of familial PTC. This result again supports the concept of considerable genetic heterogeneity in PTC.
The 8q24 locus spans from 142.70 to 154.53 cM (deCODE), which overlaps with a reported locus for autoimmune thyroid disease (AITD), including Graves disease and Hashimoto’s thyroiditis (HT) (16). It has been previously observed that PTC is more frequent in patients with HT (26–28). There is an overlap in the morphological features and molecular profiles between PTC and Hashimoto’s thyroiditis (28). Our data imply that in some subjects common susceptibility gene(s) residing in the 8q24 locus may act as predisposing gene(s) to both PTC and AITD.
Haplotypes constructed from markers in the 8q24 locus revealed a small 12 kb haplotype shared by at least 9 out of 26 PTC families. The shared haplotype is located within two known coding genes, thyroglobulin (TG) and Src-like adaptor (SLA). Thyroglobulin is the most highly expressed protein in the thyroid gland and plays important roles in thyroid function. At least 35 different loss of function germline mutations in the TG gene have been identified and linked to thyroid dyshormonogenesis, specifically autosomal recessive congenital goiter and hypothyroidism (29, 30). However it was noticed that thyroid cancer is rare in patients with TG gene mutations (31). We sequenced all of the exons and exon-intron boundary regions of the TG gene in 6 individuals carrying the shared haplotype and found no changes suggestive of being disease-causing. Taken together with our expression data there is no evidence to suggest any involvement of the TG gene in PTC predisposition. Nevertheless it is intriguing that data on genes fully contained within the TG genomic sequence suggest their involvement.
SLA is encoded by the antisense strand of introns of the TG gene, and has three isoforms. One isoform has 7 exons; the other two isoforms have 1 or 2 additional exons, respectively, functioning as 5′ UTR. Functional studies have indicated that SLA is involved in T cell-mediated immune responses (32). We sequenced all exons and exon-intron boundaries of all isoforms and did not find deleterious mutations.
In addition to the TG and SLA genes associated with the haplotype in the 8q24 locus, there are three uncharacterized cDNAs, AK023948, AK023852 and AK024366 cDNAs (33), and numerous ESTs in this genomic region. These three cDNAs reside in introns of the SLA gene; each of them is believed to be a single exon gene ranging in size from 2.1 to 2.8 kb. Although computational analysis predicts a putative open reading frame with less than 100 amino acids in each, no similarities with any known proteins seem to exist. The lack of paralogs or orthologs suggests that these transcripts are unlikely to code for proteins and may be non-coding RNA genes.
We reasoned that genes predisposing to PTC should be expressed in normal thyroid tissue and their expression level might be altered (up or down) in tumor tissue. It is noteworthy that two of the genes (AK023948 and AK023852) were originally detected in normal thyroid tissue. Expression analysis revealed that AK023948 and AK023852 are indeed abundantly expressed in normal thyroid tissue and showed significant down-expression in about 50% of the PTC of tumors we tested initially. Further, real time RT-PCR based expression analysis in 26 sporadic and 5 familial PTC patients showed downexpression of AK023948 in 24 out of 26 and 5/5 patients, respectively, often with dramatic reduction.
Given the likely low penetrance, high population frequency, and hypothetical gene-gene interaction model of PTC predisposition, the mechanism may be a subtle change in gene expression. However, in the absence of obvious deleterious mutations the basis of the reduced expression is unclear. Innocent-appearing DNA polymorphism(s) might be responsible for the reduced expression and in any case we propose that the putative non-coding RNA gene AK023948 is a candidate susceptibility gene for PTC. Taken together, we have identified a new predisposing locus in 8q24 in PTC through linkage, haplotype sharing, and gene expression analysis.
Supplementary Material
Acknowledgments
We thank K. Jazdzewski, S. Tanner, and M. Clendenning for stimulating discussions. We also thank the OSU Comprehensive Cancer Center (OSUCCC) Microarray Shared Resource for SNP genotyping; the OSUCCC Nuclei Acid Shared Resource for microsatellite marker genotyping and sequencing. Tissue samples were provided by the Cooperative Human Tissue Network which is funded by the National Cancer Institute. This work was supported by a grant P30 CA16058 and P01CA124570 from the National Cancer Institute.
Footnotes
References
- 1.Loh KC. Familial nonmedullary thyroid carcinoma: a meta-review of case series. Thyroid. 1997;7:107–13. doi: 10.1089/thy.1997.7.107. [DOI] [PubMed] [Google Scholar]
- 2.Fagin JA. Familial nonmedullary thyroid carcinoma--the case for genetic susceptibility. J Clin Endocrinol Metab. 1997;82:342–4. doi: 10.1210/jcem.82.2.3787. [DOI] [PubMed] [Google Scholar]
- 3.Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst. 1994;86:1600–8. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- 4.Dong C, Hemminki K. Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer. 2001;92:144–50. [PubMed] [Google Scholar]
- 5.Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10:733–41. [PubMed] [Google Scholar]
- 6.Frich L, Glattre E, Akslen LA. Familial occurrence of nonmedullary thyroid cancer: a population-based study of 5673 first-degree relatives of thyroid cancer patients from Norway. Cancer Epidemiol Biomarkers Prev. 2001;10:113–7. [PubMed] [Google Scholar]
- 7.Hrafnkelsson J, Tulinius H, Jonasson JG, Olafsdottir G, Sigvaldason H. Papillary thyroid carcinoma in Iceland. A study of the occurrence in families and the coexistence of other primary tumours. Acta Oncol. 1989;28:785–8. doi: 10.3109/02841868909092308. [DOI] [PubMed] [Google Scholar]
- 8.Canzian F, Amati P, Harach HR, et al. A gene predisposing to familial thyroid tumors with cell oxyphilia maps to chromosome 19p13.2. Am J Hum Genet. 1998;63:1743–8. doi: 10.1086/302164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bignell GR, Canzian F, Shayeghi M, et al. Familial nontoxic multinodular thyroid goiter locus maps to chromosome 14q but does not account for familial nonmedullary thyroid cancer. Am J Hum Genet. 1997;61:1123–30. doi: 10.1086/301610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malchoff CD, Sarfarazi M, Tendler B, et al. Papillary thyroid carcinoma associated with papillary renal neoplasia: genetic linkage analysis of a distinct heritable tumor syndrome. J Clin Endocrinol Metab. 2000;85:1758–64. doi: 10.1210/jcem.85.5.6557. [DOI] [PubMed] [Google Scholar]
- 11.McKay JD, Lesueur F, Jonard L, et al. Localization of a susceptibility gene for familial nonmedullary thyroid carcinoma to chromosome 2q21. Am J Hum Genet. 2001;69:440–6. doi: 10.1086/321979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 13.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–63. [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomer Y, Greenberg DA, Concepcion E, Ban Y, Davies TF. Thyroglobulin is a thyroid specific gene for the familial autoimmune thyroid diseases. J Clin Endocrinol Metab. 2002;87:404–7. doi: 10.1210/jcem.87.1.8291. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, Prasad M, Lemon WJ, et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. PNAS. 2001;98:15044–49. doi: 10.1073/pnas.251547398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKay JD, Thompson D, Lesueur F, et al. Evidence for interaction between the TCO and NMTC1 loci in familial non-medullary thyroid cancer. J Med Genet. 2004;41:407–12. doi: 10.1136/jmg.2003.017350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goggins W, Daniels GH, Tsao H. Elevation of thyroid cancer risk among cutaneous melanoma survivors. Int J Cancer. 2006;118:185–8. doi: 10.1002/ijc.21300. [DOI] [PubMed] [Google Scholar]
- 21.Eldon BJ, Thorlacius S, Jonsson T, et al. A population-based study on the familial aggregation of cutaneous malignant melanoma in Iceland. Eur J Cancer. 2006;42:922–6. doi: 10.1016/j.ejca.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 22.Canchola AJ, Horn-Ross PL, Purdie DM. Risk of second primary malignancies in women with papillary thyroid cancer. Am J Epidemiol. 2006;163:521–7. doi: 10.1093/aje/kwj072. [DOI] [PubMed] [Google Scholar]
- 23.Kimura ET, Nikiforova MN, Zhu Z, et al. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–7. [PubMed] [Google Scholar]
- 24.Xu X, Quiros RM, Gattuso P, Ain KB, Prinz RA. High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res. 2003;63:4561–7. [PubMed] [Google Scholar]
- 25.Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17:31–9. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Ott RA, Calandra DB, McCall A, et al. The incidence of thyroid carcinoma in patients with Hashimoto’s thyroiditis and solitary cold nodules. Surgery. 1985;98:1202–6. [PubMed] [Google Scholar]
- 27.Ott RA, McCall AR, McHenry C, et al. The incidence of thyroid carcinoma in Hashimoto’s thyroiditis. Am Surg. 1987;53:442–5. [PubMed] [Google Scholar]
- 28.Di Pasquale M, Rothstein JL, Palazzo JP. Pathologic features of Hashimoto’s-associated papillary thyroid carcinomas. Hum Pathol. 2001;32:24–30. doi: 10.1053/hupa.2001.21138. [DOI] [PubMed] [Google Scholar]
- 29.Vono-Toniolo J, Rivolta CM, Targovnik HM, Medeiros-Neto G, Kopp P. Naturally occurring mutations in the thyroglobulin gene. Thyroid. 2005;15:1021–33. doi: 10.1089/thy.2005.15.1021. [DOI] [PubMed] [Google Scholar]
- 30.Caputo M, Rivolta CM, Esperante SA, et al. Congenital hypothyroidism with goitre caused by new mutations in the thyroglobulin gene. Clin Endocrinol (Oxf) 2007;67:351–7. doi: 10.1111/j.1365-2265.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 31.Alzahrani AS, Baitei EY, Zou M, Shi Y. Metastatic follicular thyroid carcinoma arising from congenital goiter as a result of a novel splice donor site mutation in the thyroglobulin gene. J Clin Endocrinol Metab. 2006;91:740–6. doi: 10.1210/jc.2005-2302. [DOI] [PubMed] [Google Scholar]
- 32.Roche S, Alonso G, Kazlauskas A, et al. Src-like adaptor protein (Slap) is a negative regulator of mitogenesis. Curr Biol. 1998;8:975–8. doi: 10.1016/s0960-9822(98)70400-2. [DOI] [PubMed] [Google Scholar]
- 33.Ota T, Suzuki Y, Nishikawa T, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–5. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.