Abstract
Mucin 4 (MUC4) is a high molecular weight transmembrane mucin that is overexpressed in many carcinomas and is a risk factor associated with a poor prognosis. In this study, we show that the DNA methylation pattern is intimately correlated with MUC4 expression in breast, lung, pancreas and colon cancer cell lines. We mapped the DNA methylation status of 94 CpG sites from −3622 to +29 using MassARRAY analysis that utilises base-specific cleavage of nucleic acids. MUC4-negative cancer cell lines and those with low MUC4 expression (eg, A427) were highly methylated near the transcriptional start site, whereas MUC4-positive cell lines (eg, NCI-H292) had low methylation levels. Moreover, 5-aza-2′-deoxycytidine and trichostatin A treatment of MUC4-negative cells or those with low MUC4 expression caused elevation of MUC4 mRNA. Our results suggest that DNA methylation in the 5′ flanking region play an important role in MUC4 gene expression in carcinomas of various organs. An understanding of epigenetic changes in MUC4 may contribute to the diagnosis of carcinogenic risk and prediction of outcome in patients with cancer.
Keywords: MUC4 , mucin, epigenetics, DNA methylation, massARRAY
Mucins are high molecular weight glycoproteins that have oligosaccharides attached through O-glycosidic linkages to serine or threonine residues of the core protein. Many human mucin core proteins (MUC1-MUC9, MUC11-13 and MUC15-20) have been identified, (Lapensee et al, 1997; Yonezawa and Sato, 1997; Gum et al, 2002; Pallesen et al, 2002; Yin et al, 2002) including MUC4, a large transmembrane mucin with a very long glycosylated extracellular domain that is expressed in various normal tissues (Audie et al, 1993, 1995; Buisine et al, 1999; Gipson et al, 1999). MUC4 is also often overexpressed in epithelial cancers and our immunohistochemical studies have shown that aberrant expression of MUC4 is associated with invasive proliferation of tumours and a poor outcome for patients (Shibahara et al, 2004; Saitou et al, 2005; Tamada et al, 2006; Tsutsumida et al, 2007).
MUC4 also serves as a novel intramembrane ligand for the receptor tyrosine kinase ErbB2, a transmembrane glycoprotein with a tyrosine kinase domain that is encoded by the c-ErbB-2 proto-oncogene and is highly homologous with the epidermal growth factor receptor (Yamamoto et al, 1986). Furthermore, MUC4 plays an important role in cell proliferation and differentiation of epithelial cells by inducing specific phosphorylation of ErbB2 and enhancing the expression of p27 (Jepson et al, 2002), a cyclin-dependent kinase inhibitor that regulates the G1 and S phases of the cell cycle (Polyak et al, 1994).
We recently described the epigenetic regulation of the MUC1 gene (Yamada et al, 2008). Similarly to MUC1, MUC4 is a factor associated with poor prognosis in several carcinomas (Shibahara et al, 2004; Saitou et al, 2005; Tamada et al, 2006; Tsutsumida et al, 2007), and thus we hypothesised that MUC4 is also regulated by an epigenetic mechanism. To investigate the possible epigenetic regulation of MUC4 gene expression, we mapped the DNA methylation status of the MUC4 promoter region using 10 cancer cell lines derived from carcinomas of four different organs (breast, lung, pancreas and colon). Methylation of cytosine in genomic DNA plays an important role in gene regulation, and especially in gene silencing (Bird, 1992), and generally the promoter region of a transcribed gene is hypomethylated (Wolffe et al, 1999; Stirzaker et al, 2004).
To examine the methylation profiles of 92 CpG sites in the MUC4 promoter in the cancer cell lines, we performed a MassARRAY methylation analysis (Ehrich et al, 2005). On the basis of the results of this analysis, methylation-specific polymerase chain reaction (PCR; MSP) primers were designed to ensure that particular CpG sites were related to gene expression. MUC4-negative cells or those with low MUC4 expression were also treated with a DNA methylation inhibitor, 5-aza-2′-deoxycytidine, and a histone deacetylase inhibitor, trichostatin A (TSA), to confirm that DNA methylation and histone modification suppressed the expression of MUC4 mRNA. Using these results, we describe an epigenetic mechanism through which MUC4 gene expression is tightly linked to DNA methylation in several organs.
Materials and methods
Cells and treatment
Human breast cancer cell lines MCF-7 (MUC4+/−), T-47D (MUC4+/−) and MDA-MB-453 (MUC4+/−); human lung cancer cell lines NCI-H292 (MUC4+) and A427 (MUC4-); human pancreatic carcinoma cell lines HPAFII (MUC4+), BxPC3 (MUC4+) and PANC1 (MUC4+/−) and human colon adenocarcinoma cell lines LS174T (MUC4+/−) and Caco2 (MUC4+/−) were obtained from American Type Culture Collection (Manassas, VA, USA). MCF-7, A427, HPAFII, Caco2 and LS174T cells were cultured in Eagle’s minimum essential medium (Sigma, St Louis, MO, USA); PANC1 cells were cultured in D-MEM (Sigma); T-47D, NCI-H292 and BxPC3 cells were cultured in RPMI-1640 medium (Sigma) and MDA-MB-453 cells were cultured in Leibovitz’s L-15 medium (Invitrogen, Carlsbad, CA, USA). All media were supplemented with 10% foetal bovine serum (Invitrogen) and 100 U ml−1 penicillin 100 μg ml−1 streptomycin (Sigma). MUC4-negative cells or cells with low MUC4 expression were split 24 h before treatment. MDA-MB-453, PANC1 and LS174T cells were incubated with 100 μM 5-aza-2′-deoxycytidine (5-AzadC; Sigma) and/or 500 nM TSA(Sigma) for 5 days, MCF-7, T-47D and Caco2 cells were incubated with 100 μM 5-AzadC and/or 50 nM TSA for 5 days, and A427 cells were incubated with 1 μM 5-AzadC and/or 50 nM TSA for 5 days. Media were changed every 24 h.
Quantitative reverse transcription PCR (RT—PCR) analysis
Messenger RNA from cells that had or had not been treated with 5-AzadC, TSA or 5-AzadC/TSA in combination was purified with an RNeasy Mini kit (Qiagen, Valencia, CA, USA). Of a total of 100 μl of mRNA, 20 μl was reverse transcribed with Random-Hexamers (Applied Biosystems, Foster City, CA, USA). A 3.5-μl cDNA aliquot was amplified in 25 μl of 2xTaqMan universal master mix, 2.5 μl of 20xTarget assay mix and 2.5 μl of 20x control assay mix (Applied Biosystems) under the following PCR conditions: 2 min at 50°C, 10 min at 95°C, 45 cycles for 15 s at 95°C and 1 min at 60°C. The primers and probes were designed and synthesised by Applied Biosystems. The Target assay mix used for MUC4 had the product number Hs003666414. Human GAPDH (product number 4310884E) was used to calibrate the original concentration of mRNA; that is, the concentration of mRNA in the cell was defined as the ratio of target mRNA copies relative to GAPDH mRNA copies. In this analysis, data from three separate experiments were averaged.
MUC4 gene promoter sequencing
Genomic DNA was extracted from the 10 cell lines using a DNeasy tissue system (Qiagen) according to the manufacturer’s instructions. DNA was PCR amplified using seven pairs of sense and antisense primers (Table 1) for the full-length MUC4 promoter. Polymerase chain reaction fragments were sequenced using a single-strand sequencing method (Hokkaido System Science Co., Hokkaido, Japan). Sequences were analysed with an ABI Prism 310 Genetic Analyzer (PE Applied Biosystems).
Table 1. Synthetic oligonucleotides used in the study.
| Name | Primer sequence | Position |
|---|---|---|
| MUC4 promoter sequencing primers | ||
| MUC4-S1L | AGGCAGAACAGAGCTCAAATTC | −3647 to −3626 |
| MUC4-S1R | ACAAAATGAGTGGCTTCTCTAGTTC | −2970 to −2946 |
| MUC4-S2L | ACTTACACCTGACTTCAGACTCCTG | −3047 to −3023 |
| MUC4-S2R | TTCCTCGTTCACCTGTAAAATG | −2,410 to −2389 |
| MUC4-S3L | AACCAGGTGTCGTAACAACAAG | −2523 to −2504 |
| MUC4-S3R | ACCATCTCAGCTCACTGCAAG | −1846 to −1826 |
| MUC4-S4L | AAGAGCTGACAGAAAAATGACCAC | −2,092 to −2,069 |
| MUC4-S4R | TGTAGCTCTCTGCAAAGGAAAAC | −1236 to −1214 |
| MUC4-S5L | CCATGAACAGACTTTCAGCTTCTAG | −1521 to −1497 |
| MUC4-S5R | CCAGATGGTGGACTTCTTCTTTC | −958 to −936 |
| MUC4-S6L | CCTACCGAGGTTTTGCCTTC | −1063 to −1044 |
| MUC4-S6R | CTAATCACCCTTTTCTCTCCTCAG | −194 to −171 |
| MUC4-S7F | CATACAGTGCATTTCTGTTCCTG | −557 to −535 |
| MUC4-S7R | ATCACTTACCTGGGACCACATG | +70 to +91 |
| 10mer-tagged or T7-tagged primers a | ||
| MUC4-1L | aggaagagagTGTGTGGTTTAGAAGGTTGTAATTG | −3611 to −3587 |
| MUC4-1R | cagtaatacgactcactatagggagaaggctACCCAAATAAAAATCCCTAATTTTT | −3233 to −3208 |
| MUC4-2L | aggaagagagTGGGAGAATTTTATGATTTTATTTTTGTT | −3134 to −3106 |
| MUC4-2R | cagtaatacgactcactatagggagaaggctATTTCCAAACCCAAATCTTTCCTAC | −2671 to −2647 |
| MUC4-3L | aggaagagagGTAGGAAAGATTTGGGTTTGGAAAT | −2671 to −2647 |
| MUC4-3R | cagtaatacgactcactatagggagaaggctAACATCCAAATAAAACCAAACTAAAC | −2204 to −2179 |
| MUC4-4L | aggaagagagTGTGTGTTTTTGGTTTTTTTGGTAT | −2274 to −2250 |
| MUC4-4R | cagtaatacgactcactatagggagaaggctACACCATCTCAACTCACTACAAACTC | −1849 to −1824 |
| MUC4-5L | aggaagagagTAGTTTTTTGTTTTGGAAAGAAGAAGTTTA | −973 to −944 |
| MUC4-5R | cagtaatacgactcactatagggagaaggctAACAAACCCCAAAAAATTAAAAAAC | −512 to −488 |
| MUC4-6L | aggaagagagGGTTTGTTATGTGTTTGGGATTTG | −793 to −770 |
| MUC4-6R | cagtaatacgactcactatagggagaaggctAAAATAAATAAACCACCCTCCTAAC | −344 to −320 |
| MUC4-7Lb | aggaagagagTAGGGATATTTAGGGGATTTTTTTT | +18 to +42 |
| MUC4-7Rb | cagtaatacgactcactatagggagaaggctTTCCCAACAACCCAAAACTCTAATA | −415 to −391 |
| MSP primers | ||
| MSP-ULc | GGTGATTAGTGTGGGGTTTTG | −179 to −159 |
| MSP-UR | CCAAACCAAATACATTTCTCCAA | −122 to −100 |
| MSP-MLd | GGTGATTAGCGTGGGGTTTC | −179 to −160 |
| MSP-MR | CGAACCAAATACGTTTCTCCG | −121 to −101 |
L=(aggaagagag) + (gene-specific sequence), R=(cagtaatacgactcactatagggagaaggct) + (gene-specific sequence).
MUC4-7L and MUC4-7R primers were designed for the antisense strand because PCR amplification was not obtained in the forward direction.
U indicates the primer for unmethylated alleles.
M indicates the primer for methylated alleles.
Quantitative methylation analysis
Quantitative methylation analysis of the MUC4 promoter was performed using the MassARRAY compact system (Hitachi high technologies corporation, Tokyo, Japan; Ehrich et al, 2005; Yamada et al, 2008). DNA from the cell lines was extracted using a DNeasy tissue system (Qiagen). A 1-mg sample of DNA was converted with sodium bisulfite using an EZ DNA methylation kit (Zymo research, Orange, CA, USA) and the modified DNA was amplified by PCR. The target regions were amplified using the primer pairs shown in Table 1. Each forward primer was tagged with a 10mer (5′-AGGAAGAGAG-3′) to balance the PCR, and each reverse primer had a T7-promoter tag (5′-CAGTAATACGACTCACTATAGGGAGAAGGCT-3′) for in vitro transcription. Polymerase chain reaction amplification was performed with the following parameters: hot start at 94°C for 15 min, followed by denaturing at 94°C for 20 s, annealing at 56°C for 30 s, extension at 72°C for 1 min for 45 cycles and final incubation at 72°C for 3 min. Unincorporated dNTPs were dephosphorylated by adding 2 μl of premix including 0.3 U shrimp alkaline phosphate (SAP; Sequenom, San Diego, CA, USA). The reaction mixture was incubated at 37°C for 40 min and SAP was then heat inactivated for 5 min at 85°C. After SAP treatment, 2 μl of the PCR products were used as a template for in vitro transcription, and RNase A cleavage was used for the reverse reaction, following the manufacturer's instructions (Sequenom). The samples were conditioned and spotted on a 384-pad Spectro-CHIP (Sequenom) using a MassARRAY nanodispenser (Samsung, Irvine, CA, USA), followed by spectral acquisition on a MassARRAY analyzer compact MALDI-TOF MS (Sequenom). The resultant methylation calls were analysed with EpiTyper software v1.0 (Sequenom) to generate quantitative results for each CpG site or an aggregate of multiple CpG sites.
DNA extraction and DNA MSP analysis
DNA from the cell lines was extracted using a DNeasy tissue system (Qiagen) according to the manufacturer’s instructions. Bisulfite modification of the genomic DNA was carried out using a Epitect Bisulfite Kit (Qiagen), and the modified DNA was amplified by PCR using a AmpliTaq Gold Fast PCR Kit (Applied Biosystems). The target regions were amplified using the primer pairs shown in Table 1. The PCR conditions were 95°C for 10 min, 40 cycles at 96°C for 3 s, 59°C for 3 s and 68°C for 3 s, with a final extension reaction at 72°C for 10 s. The amplified products were subjected to 1.5% agarose gel electrophoresis.
Results
Effects of 5-AzadC and TSA on MUC4 mRNA expression
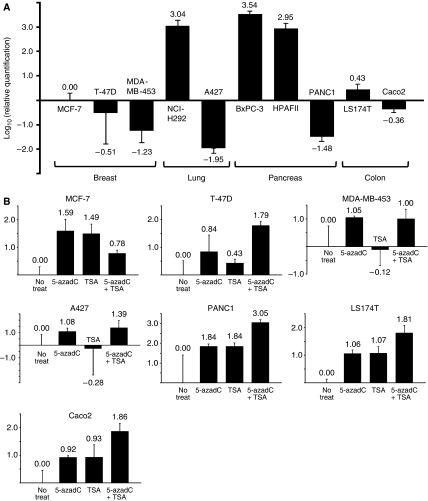
The expression levels of MUC4 mRNA in the human breast cancer cell lines MCF-7, T-47D and MDA-MB-453, the human lung cancer cell lines NCI-H292 and A427, the human pancreatic carcinoma cell lines HPAFII, BxPC-3 and PANC1 and the human colon adenocarcinoma cell lines LS174T and Caco2 were examined using quantitative RT–PCR analysis (Figure 1A). NCI-H292, BxPC-3 and HPAFII cells expressed MUC4 mRNA, but MCF-7, T-47D, MDA-MB-453, A427, PANC1, LS174T and Caco2 cells did not do so. To examine whether DNA methylation and histone modification suppress the MUC4 mRNA expression level, MUC4-negative cells or cells with low expression of MUC4 were treated with a DNA demethylating agent, 5-AzadC, a histone deacetylase inhibitor, TSA or 5-AzadC/TSA in combination (Figure 1B). Quantitative RT–PCR analysis of MCF-7 breast cancer cells showed that treatment with 5-AzadC or TSA gave a greater increase in MUC4 mRNA compared to treatment with 5-AzadC/TSA in combination (38.9-, 30.9- and 6.03-fold increases, respectively). In contrast, in T-47D cells, the combination treatment was more effective than 5-AzadC or TSA alone in increasing MUC4 mRNA (61.7-, 6.92- and 2.69-fold increases, respectively). Following 5-AzadC or 5-AzadC/TSA treatment of MDA-MB-453 cells, MUC4 mRNA expression increased by 11.2- and 10.0-fold, respectively, but TSA alone had no effect. Treatment of A427 cells with 5-AzadC or 5-AzadC/TSA resulted in an increase in MUC4 mRNA of 12.0- to 24.5-fold, whereas the level remained constant after treatment with TSA alone. In PANC1, LS174T and Caco2 cells, treatment with 5-AzadC or TSA increased the MUC4 mRNA levels and 5-AzadC/TSA in combination further increased these levels (by 69.2-, 69.2- and 1,120-fold, respectively, in PANC1 cells; 11.5-, 11.7- and 64.6-fold, respectively, in LS174T cells; and 8.32-, 8.51- and 72.4-fold, respectively, in Caco2 cells). These results suggest that DNA methylation and/or histone modification influence MUC4 gene expression.
Figure 1.
Quantitative RT–PCR analysis of 10 cancer cell lines before and after treatment. (A) Expression of MUC4 mRNA examined by quantitative RT–PCR. The bar graphs show gene expression levels relative to those in MCF-7 cells. NCI-H292, BxPC-3 and HPAFII cells showed high expression of MUC4 mRNA, whereas MCF-7, T-47D, MDA-MB-453, A427, PANC1, LS174T and Caco2 cells had no or low levels of MUC4 mRNA. (B) Quantitative analysis of MUC4 mRNA in cells with little or no MUC4 expression after treatment with 5-AzadC, TSA and 5-AzadC/TSA in combination. The bar graphs show gene expression levels as log10 values relative to those in untreated cells. After 5-AzadC or TSA treatment, MCF-7 cells showed significant restoration of MUC4 mRNA expression. In T-47D cells, 5-AzadC/TSA treatment was most effective in restoring expression. MUC4 mRNA levels were markedly restored in MDA-MB-453 and A427 cells after treatment with 5-AzadC or 5-AzadC/TSA. In PANC1, LS174T and Caco2 cells, 5-AzadC/TSA treatment was more effective in restoring expression compared to treatment with 5-AzadC or TSA alone.
DNA sequencing of the human MUC4 mucin gene promoter
To investigate whether MUC4 gene expression is regulated by SNPs, sequencing primers were designed to target different regions of the MUC4 promoter (Table 1) based on the published sequence (Perrais et al, 2001). We sequenced a 3.6-kb promoter region of the human MUC4 from 10 cell lines (Figure 2) and found that bases at some positions differed among the 10 cell lines. However, there was no correlation between these sequence differences and MUC4 gene expression.
Figure 2.
The human MUC4 gene promoter sequence, which spans positions −3,622 to +29 with respect to the transcription start site. The numbers of CpG sites and the transcriptional start site +1 (arrow) are shown. CpG sites 53–76 were undetectable by MassARRAY analysis in the 10 cell lines. Between bases −833 to −648, sequence variation was observed among the 10 cell lines.
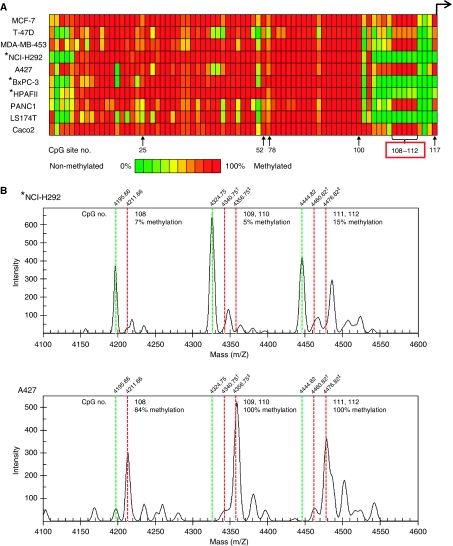
Quantitative MassARRAY methylation analysis of the MUC4 gene promoter in 10 cancer cell lines
Quantification of promoter DNA methylation levels was performed using the MassARRAY compact system, with mapping of the resulting data (Figure 3A). Samples were analysed by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF-MS), which permits high-throughput identification of methylation sites and semiquantitative measurement at single or multiple CpG sites. The raw spectrum data for CpG sites 108–112 in NCI-H292 and A427 lung cancer cells are shown in Figure 3B. In the vicinity of the transcriptional start site, MUC4-negative cells or cells with low expression of MUC4 showed high methylation (MCF-7, T-47D, MDA-MB-453, A427, PANC1 and Caco2) compared with MUC4-positive cell lines (NCI-H292, BxPC-3 and HPAFII). In contrast, the MUC4 promoter CpG sites were mostly hypomethylated in LS174T cells, which showed low expression of MUC4. In the 10 cancer cell lines, methylation of CpG sites 108–112 was inversely correlated with MUC4 gene expression, whereas methylation of sites 1–107 was almost unrelated to gene expression. These results indicate that the CpG methylation status near to the MUC4 transcriptional start site may play an important role in gene expression of MUC4.
Figure 3.
Summary of the CpG methylation status of the MUC4 gene promoter region in 10 cancer cell lines. (A) Quantitative methylation analysis of CpG sites located in the MUC4 promoter using a MassARRAY compact system. Different colours display relative methylation changes in 10% increments (green=0%, red=100% methylated). CpG sites that correlated well with MUC4 expression are boxed in red. (B) Raw spectrum data for CpG sites 108–112 in lung cancer cell lines. Non-methylated and methylated CpG sites generate status-specific mass signals. Methylation status (%) is calculated as (intensity of methylated CpG (red dotted line)/(intensity of methylated CpG (red dotted line)+intensity of unmethylated CpG (green dotted line))). † Indicates methylation in one of the two CpG sites (eg, 109 or 110). ‡ Indicates methylation of both CpG sites (eg, 109 and 110). Asterisks indicate MUC4-positive cell lines.
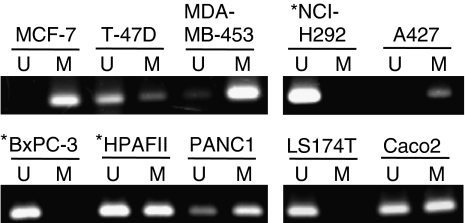
Methylation-specific PCR analysis of MUC4 promoter methylation in 10 cancer cell lines
Methylation-specific PCR primers were designed to target CpG sites in the 5′ flanking region of the MUC4 promoter (Table 1) based on the results of MassARRAY analysis. The five CpG sites (Nos. 108–112) included in the MSP primer showed 4–39% methylation in MUC4-positive cells and 63–100% methylation in all MUC4-negative/low cells, except for LS174T cells. The LS174T cells showed 5–19% methylation at the five sites. To confirm the reliability of the MSP primer, MSP analysis was performed on the 10 cell lines (Figure 4). An unmethylated band (lanes indicated by U in Figure 4) was obtained in MUC4-positive NCI-H292 and BxPC-3 cells, whereas a methylation band (lanes indicated by M in Figure 4) was observed in MUC4-negative/low MCF-7, MDA-MB-453, A427 and PANC1 cells. The MSP data were almost consistent with the results of MassARRAY analysis of the methylation status of the cell lines. Therefore, our results suggest that the 5′ flanking region of the promoter, and especially CpG sites 108–112, may play an important role in methylation-related gene silencing of MUC4.
Figure 4.
Methylation-specific PCR (MSP) analysis of the MUC4 gene promoter region in 10 cancer cell lines. The PCR products labelled as ‘M’ (methylated) were generated by methylation-specific primers and those labelled as ‘U’ (unmethylated) were generated by primers specific for unmethylated DNA. The unmethylated allele was detected in MUC4-positive NCI-H292 and BxPC-3 cells, whereas the methylated allele was detected in MUC4-negative/low MCF-7, MDA-MB-453, A427 and PANC1 cells. Asterisks indicate MUC4-positive cell lines.
Discussion
We have previously examined the expression profiles of MUC4 in primary tumours of patients with intrahepatic cholangiocarcinoma-mass forming type (n=27; Shibahara et al, 2004), pancreatic adenocarcinoma (n=135) (Shibahara et al, 2004; Saitou et al, 2005; Tamada et al, 2006; Tsutsumida et al, 2007), extrahepatic bile duct carcinoma (n=70; Tamada et al, 2006) and small-sized lung adenocarcinoma (less than 3 cm; n=185; Tsutsumida et al, 2007) and compared MUC4 expression with the survival of the patients. In the four series, survival in MUC4-positive or high-expression cases was significantly worse than that for MUC4-negative or low-expression cases, and multivariate analyses showed that MUC4 expression in the carcinomas was a risk factor for a poor prognosis (Shibahara et al, 2004; Saitou et al, 2005; Tamada et al, 2006; Tsutsumida et al, 2007).
The regulatory mechanism of the MUC4 gene is unclear. Initially, to examine the possible epigenetic regulation of MUC4 expression, we treated MUC4-negative or low-expression cancer cell lines with 5-AzadC and/or TSA (Figure 1B). In four of seven cells (T-47D, PANC1, LS174T and Caco2), treatment with 5-AzadC/TSA significantly restored the MUC4 mRNA expression, compared to the treatment with 5-AzadC or TSA alone. In MCF-7 cells, however, the restoration level of MUC4 mRNA decreased in 5-AzadC/TSA treatments rather than in treatment with 5-AzadC or TSA alone. Yet, it is unclear whether 5-AzadC and TSA caused an antagonistic effect in MCF-7 cells. We showed that treatment with TSA did not restore the MUC4 mRNA in MDA-MB-453 and A427 cells. Kondo et al (2003) showed that 5-AzadC or a combination of 5-AzadC and TSA, but not TSA alone, reactivates tumour suppressor gene expression at silenced loci (eg, p16). Our results in MDA-MB-453 and A427 cells are in good agreement with their observation. Although there were differences in the restoration level of MUC4 mRNA among treated cell lines, our overall results suggested the possibility that MUC4 expression is regulated by epigenetic mechanisms.
Next, to rule out the contribution of SNPs in control of MUC4 expression, the sequences of the MUC4 promoter were verified in all the cells used in the study using primers based on the published sequence (Perrais et al, 2001). In Figure 2, the sequence of part of the promoter (−833 to −647) could not be determined by single-strand sequencing, and we were also unable to obtain reliable results for one promoter region (−1822 to −973) using the MassARRAY compact system. Moreover, we found that bases at some positions differed among the 10 cell lines in a 3.6-kb MUC4 promoter region. However, we found no correlation between these sequence differences and MUC4 expression, and therefore we examined the DNA methylation status of the MUC4 promoter in 10 cancer cell lines.
In this study, we used MassARRAY analysis to examine the methylation status of 92 CpG sites in the promoter of the MUC4 gene. Methylation at CpG sites 108–112 (−170 to −102) was associated with the expression of MUC4 in all cell lines except for LS174T. MUC4-negative cell lines were hypermethylated at these five CpG sites, whereas MUC4-positive cell lines showed hypomethylation at the same sites. We recently reported that MUC1 expression in pancreatic, breast and colon cancer cells is regulated by DNA methylation and histone H3-K9 modification in the 5′ flanking region, also using a MassARRAY compact system. Methylation of CpG near the transcriptional start site (−100 to 100) was inversely correlated with MUC1 gene expression (Yamada et al, 2008). Similarly, MUC4 expression was also regulated by methylation of CpG sites near the transcriptional start site (−170 to −102).
On the other hand, the methylation status of upstream regions clearly differed between MUC1 and MUC4. In MUC1, most CpG sites except for the regulatory region were unmethylated (Yamada et al, 2008); in contrast, in MUC4, almost all CpG sites of the upstream region (until CpG site 100) were methylated (Figure 3A). The reason for the different global methylation levels of these similar transmembrane mucins requires further investigation.
We were unable to show a relationship between DNA methylation status and expression of the MUC4 gene in the LS174T colon cancer cell line, similarly to our results for MUC1 (Yamada et al, 2008). However, expression of MUC4 in Caco2 cells, which are derived from cancer cells of the same organ, was related to the DNA methylation status. This indicates that the regulation of expression by DNA methylation is not organ-specific, but varies in individual cell lines.
Recently, Vincent et al (2008) reported that MUC4 expression is regulated epigenetically through DNA methylation and histone modification in pancreatic and gastric epithelial cancer cell lines, and that CpG sites 110–114 (−121 to −81) were associated with expression of MUC4 in these cells (Vincent et al, 2008). However, in our study, CpG 113 (−93) and 114 (−81) were hypomethylated in all cell lines and unrelated to the expression of MUC4 gene, whereas sites 108 and 109 were associated with MUC4 gene expression. These differences might be due to different methods of detection, and it also possible that these CpG sites (108, 109, 113 and 114) may be sensitive and unstable. This issue may be resolved by the use of different analytical methods in future studies.
MUC4 is a factor associated with a poor prognosis in various carcinomas and particularly in intractable carcinomas, such as pancreatobiliary and lung carcinomas (Saitou et al, 2005; Tamada et al, 2006; Tsutsumida et al, 2007). This study shows that MUC4 expression is regulated by DNA methylation in the promoter region, and we have also shown that MUC1, another prognostic indicator, is regulated by a similar epigenetic mechanism (Yamada et al, 2008). These results indicate that MassARRAY is a powerful tool for detection of methylation of CpG sites, and screening of the DNA methylation status of MUC1 and MUC4 genes by MassARRAY analysis of discharged fluids, such as pancreatic juice, bile or sputum is likely to be of value for early detection of pancreatobiliary and lung carcinomas.
Acknowledgments
We thank Mr Hiromichi Iai (Hitachi High-Technologies Corporation, Tokyo, Japan) and Mr Mitsuhiro Miyazaki (Hitachi High-Tech Manufacturing and Service Corporation, Ibaraki, Japan) for their help with quantitative DNA methylation analysis. This work was supported in part by a grant-in-aid from the Ministry of Education, Science, Sports, Culture and Technology, Japan (Scientific Research on Priority Areas 18014024 and 20014022, and Scientific Research (C) 20590345 to S Yonezawa).
References
- Audie JP, Janin A, Porchet N, Copin MC, Gosselin B, Aubert JP (1993) Expression of human mucin genes in respiratory, digestive, and reproductive tracts ascertained by in situ hybridization. J Histochem Cytochem 41: 1479–1485 [DOI] [PubMed] [Google Scholar]
- Audie JP, Tetaert D, Pigny P, Buisine MP, Janin A, Aubert JP, Porchet N, Boersma A (1995) Mucin gene expression in the human endocervix. Hum Reprod 10: 98–102 [DOI] [PubMed] [Google Scholar]
- Bird A (1992) The essentials of DNA methylation. Cell 70: 5–8 [DOI] [PubMed] [Google Scholar]
- Buisine MP, Devisme L, Copin MC, Durand-Reville M, Gosselin B, Aubert JP, Porchet N (1999) Developmental mucin gene expression in the human respiratory tract. Am J Respir Cell Mol Biol 20: 209–218 [DOI] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D (2005) Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA 102: 15785–15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson IK, Spurr-Michaud S, Moccia R, Zhan Q, Toribara N, Ho SB, Gargiulo AR, Hill III JA (1999) MUC4 and MUC5B transcripts are the prevalent mucin messenger ribonucleic acids of the human endocervix. Biol Reprod 60: 58–64 [DOI] [PubMed] [Google Scholar]
- Gum Jr JR, Crawley SC, Hicks JW, Szymkowski DE, Kim YS (2002) MUC17, a novel membrane-tethered mucin. Biochem Biophys Res Commun 291: 466–475 [DOI] [PubMed] [Google Scholar]
- Jepson S, Komatsu M, Haq B, Arango ME, Huang D, Carraway CA, Carraway KL (2002) Muc4/sialomucin complex, the intramembrane ErbB2 ligand, induces specific phosphorylation of ErbB2 and enhances expression of p27(kip), but does not activate mitogen-activated kinase or protein kinaseB/Akt pathways. Oncogene 21: 7524–7532 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Issa JP (2003) Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol 23: 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapensee L, Paquette Y, Bleau G (1997) Allelic polymorphism and chromosomal localization of the human oviductin gene (MUC9). Fertil Steril 68: 702–708 [DOI] [PubMed] [Google Scholar]
- Pallesen LT, Berglund L, Rasmussen LK, Petersen TE, Rasmussen JT (2002) Isolation and characterization of MUC15, a novel cell membrane-associated mucin. Eur J Biochem 269: 2755–2763 [DOI] [PubMed] [Google Scholar]
- Perrais M, Pigny P, Ducourouble MP, Petitprez D, Porchet N, Aubert JP, Van Seuningen I (2001) Characterization of human mucin gene MUC4 promoter: importance of growth factors and proinflammatory cytokines for its regulation in pancreatic cancer cells. J Biol Chem 276: 30923–30933 [DOI] [PubMed] [Google Scholar]
- Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A (1994) p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Saitou M, Goto M, Horinouchi M, Tamada S, Nagata K, Hamada T, Osako M, Takao S, Batra SK, Aikou T, Imai K, Yonezawa S (2005) MUC4 expression is a novel prognostic factor in patients with invasive ductal carcinoma of the pancreas. J Clin Pathol 58: 845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara H, Tamada S, Higashi M, Goto M, Batra SK, Hollingsworth MA, Imai K, Yonezawa S (2004) MUC4 is a novel prognostic factor of intrahepatic cholangiocarcinoma-mass forming type. Hepatology 39: 220–229 [DOI] [PubMed] [Google Scholar]
- Stirzaker C, Song JZ, Davidson B, Clark SJ (2004) Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res 64: 3871–3877 [DOI] [PubMed] [Google Scholar]
- Tamada S, Shibahara H, Higashi M, Goto M, Batra SK, Imai K, Yonezawa S (2006) MUC4 is a novel prognostic factor of extrahepatic bile duct carcinoma. Clin Cancer Res 12: 4257–4264 [DOI] [PubMed] [Google Scholar]
- Tsutsumida H, Goto M, Kitajima S, Kubota I, Hirotsu Y, Wakimoto J, Batra SK, Imai K, Yonezawa S (2007) MUC4 expression correlates with poor prognosis in small-sized lung adenocarcinoma. Lung Cancer 55: 195–203 [DOI] [PubMed] [Google Scholar]
- Vincent A, Ducourouble MP, Van Seuningen I (2008) Epigenetic regulation of the human mucin gene MUC4 in epithelial cancer cell lines involves both DNA methylation and histone modifications mediated by DNA methyltransferases and histone deacetylases. FASEB J 22: 3035–3045 [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Jones PL, Wade PA (1999) DNA demethylation. Proc Natl Acad Sci USA 96: 5894–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N, Nishida Y, Tsutsumida H, Hamada T, Goto M, Higashi M, Nomoto M, Yonezawa S (2008) MUC1 expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Res 68: 2708–2716 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Ikawa S, Akiyama T, Semba K, Nomura N, Miyajima N, Saito T, Toyoshima K (1986) Similarity of protein encoded by the human c-erb-B-2 gene to epidermal growth factor receptor. Nature 319: 230–234 [DOI] [PubMed] [Google Scholar]
- Yin BW, Dnistrian A, Lloyd KO (2002) Ovarian cancer antigen CA125 is encoded by the MUC16 mucin gene. Int J Cancer 98: 737–740 [DOI] [PubMed] [Google Scholar]
- Yonezawa S, Sato E (1997) Expression of mucin antigens in human cancers and its relationship with malignancy potential. Pathol Int 47: 813–830 [DOI] [PubMed] [Google Scholar]