Introduction
The genomes of eukaryotes are packaged into chromatin, the fundamental unit of which is the nucleosome, composed of a histone octamer. The position of the nucleosomes and the chemical modifications of histones are critical to a large number of cellular functions, including changes in gene expression during cell differentiation. Features of chromatin structure and histone modifications are also involved in the control of genome stability, both to faithfully perpetuate chromosomal organization and to dynamically regulate chromatin structure to allow DNA repair.
One of the most studied histone modifications is histone H3 lysine 4 (H3K4) methylation (Dehe and Geli, 2006; Ruthenburg et al, 2007). In vertebrates, H3K4 di- and trimethylation (H3K4me2 and H3K4me3) occur in discrete zones in the proximity of transcriptionally active gene promoters (Schneider et al, 2004; Bernstein et al, 2005; Barski et al, 2007; Heintzman et al, 2007). Genome-wide studies in exponentially growing Saccharomyces cerevisiae cells show that H3K4me3 peaks at the start of transcribed portions of genes (Pokholok et al, 2005). H3K4me3 is thought to facilitate transcription through the recruitment of nucleosome remodelling complexes and histone-modifying enzymes, and by preventing repressors from binding to chromatin (Berger, 2007; Venkatasubrahmanyam et al, 2007). However, the function of H3K4me3 in gene activation is not clear, as the expression of most genes was found to be unchanged in the absence of H3K4 methylation (Bernstein et al, 2002). Besides transcription, the H3K4me3 mark is associated with other biological functions. H3K4me3 has an important function in mammalian V(D)J recombination by recruiting the RAG2 protein through recognition of its PHD domain (Liu et al, 2007; Matthews et al, 2007). In this study, we have uncovered the essential function of H3K4me3 in the initiation of meiotic recombination, distinct from its tight association with meiotic gene expression.
During meiosis, a diploid cell produces haploid gametes (spores in yeast) for sexual reproduction, with two consecutive rounds of chromosomes segregation. Unique chromosomal events occur during MI prophase (Zickler and Kleckner, 1999), including genome-wide homologous recombination, initiated by programmed formation of double-strand breaks (DSBs) and essential for proper chromosome segregation and fertility (Hassold et al, 2007). Meiosis also involves substantial transcriptional reprogramming (Chu et al, 1998; Primig et al, 2000). Both massive changes of expression and induction of recombination are expected to involve chromatin structure modifications.
A first hint for a function of chromatin structure in DSB formation was the finding that the recombination hot spots are located in chromatin that is constitutively open before entry into meiosis (Ohta et al, 1994; Wu and Lichten, 1994) and exhibit a specific increase of sensitivity to MNase, shortly before DSB formation (Ohta et al, 1994; Murakami et al, 2003). In addition, DSB formation at well-characterized hot spots is strongly reduced in the absence of Set1, the only H3K4 methyltransferase in S. cerevisiae (Sollier et al, 2004). Finally, Rad6, which mediates methylation of H3K4 through ubiquitylation of the H2B lysine 123, is also required for full levels of meiotic DSBs (Yamashita et al, 2004). These and other studies on S. cerevisiae (Mieczkowski et al, 2007), S. pombe (Yamada et al, 2004) and Caenorhabditis elegans (Reddy and Villeneuve, 2004) suggest that post-translational histone modifications regulate DSB formation, but histone states at or near the DSB sites remain to be examined and the local versus general features of the DSB regions remain to be distinguished.
Here, we have studied in S. cerevisiae the relations between H3K4me3 levels and both meiotic mRNA levels and DSB formation. We show that the level of H3K4me3 is constitutively high in DSB-prone regions, independently of local gene transcript level, and that without Set1, DSB formation is severely reduced at 84% of the wild-type DSB sites, the reduction being quantitatively correlated with the level of H3K4me3 in wild-type cells. We conclude that H3K4me3 is a prominent mark of active meiotic recombination initiation sites. Our data also provide new insights to explain the heterogeneous distribution of recombination initiation events along the chromosomes as well as the evolution of recombination initiation sites without major DNA sequence modification.
Results
Meiotic DSBs are strongly reduced in the absence of H3K4 methylation
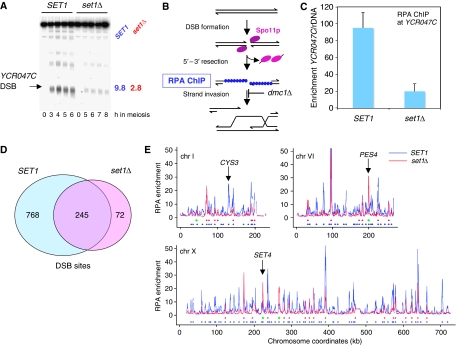
As DSB frequencies are reduced about five-fold at the YCR047C and CYS3 loci in set1Δ cells (Sollier et al, 2004) (Figure 1A), we asked to which extent Set1 controls DSB formation in the entire genome. To determine the distribution of DSB frequencies in SET1 and set1Δ cells, we used cells mutated for DMC1, which encodes the meiosis-specific Rad51 homologue. In such cells, all meiotic DSBs are formed, but accumulate with 3′ ended single strand tails, covered with replication protein A (RPA). At the time when DSBs accumulate (5 h in the SET1 and 7 h in the set1Δ cells), we performed chromatin immunoprecipitation of Rfa1, an RPA component (Johnson et al, 2007) (Figure 1B). Quantitative PCR analysis of immunoprecipitated DNA confirmed the enrichment at the DSB hot spot (YCR047C) fragment relative to ribosomal RNA genes, where meiotic DSBs are absent (Figure 1C). The enrichment was significant in both strains, but five times lower in the set1Δ strain. This is identical to the reduction of DSB frequency observed by Southern blot at this locus (Sollier et al, 2004 and Figure 1A). It has been shown recently that meiotic DSBs occur at many places in the genome, but with varying frequencies (Buhler et al, 2007). The genome-wide mapping of the DSB sites detected as RPA-enriched sites by ChIP-chip (Supplementary Figure S1 and Table S1) allowed us to define the ‘hottest' DSB sites in the genome, for both WT and set1Δ strains, as having a five-fold enrichment over experimental background. In the SET1 strain, there were 1013 such hot spots, but only 317 in the set1Δ strain (Figure 1D and Supplementary Tables S2 and S3). Out of the 1013 hottest DSBs, 742 (73%) are located at less than 1 kb from one of the hottest dmc1Δ DSBs mapped by a different approach but defined with similar criteria (Buhler et al, 2007), and out of the 100 strongest DSBs, 95 DSB sites are found also in Buhler's study, showing that the results from the two studies are very similar (see Supplementary Figure S2 for comparison of our set of DSBs with that of Buhler et al. and of Blitzblau et al, 2007). A total of 84% of the DSB hot spots of the wild-type strain show a greater than 1.5-fold reduction in the absence of Set1 (70 % exhibit a more than two-fold reduction) and the majority (77%) of the DSB hot spot peaks in set1Δ coincides with the DSB hot spot peaks observed in wild type (Figure 1D). However, even if globally the DSB profiles are flatter in the absence of Set1, locus-specific differences exist, such as that on chromosomes VI and X near the PES4 and SET4 loci, respectively, which show stronger DSB in set1Δ (Figure 1E). Further analyses of DSB formation in the absence of Set1 are presented later. In summary, our results show that with few exceptions, the inactivation of Set1 severely reduces DSB frequencies. These results led us to examine the genome-wide distribution of H3K4 trimethylation mark during meiosis.
Figure 1.
Meiotic DSB formation is globally reduced in the absence of histone H3K4 methylation. (A) Southern blot analysis of DSB formation in the promoter of YCR047C (arrow). The blot contains AseI-digested DNA from meiotic samples of SET1 dmc1Δ (ORD7354) and set1Δ dmc1Δ (ORD9624) cells. Maximal DSB frequencies (% total lane signal) observed during each time course are indicated to the right of the blot. (B) Schematic representation of the procedure using ChIP of RPA to enrich for meiotic DSBs accumulating in dmc1Δ cells. (C) Quantitative PCR measurement of RPA enrichment for sequences close to the YCR047C DSB hot spot relative to ribosomal DNA (rDNA). Data are from two independent samples for each strain taken at 5 h (SET1 dmc1Δ) or 7 h (set1Δ dmc1Δ) in meiosis. Error bars represent standard deviations. (D) The number of DSB sites is strongly reduced in the absence of SET1. The Venn diagram shows the overlap between the DSBs sites occurring in dmc1Δ and in set1Δ dmc1Δ. The DSB peaks were identified from the RPA chip-chip data after smoothing (Supplementary data). (E) Examples of DSBs signals in set1Δ. Three chromosomes are represented. For each one, unsmoothed array signals of RPA enrichment over background are displayed according to their chromosomal coordinates. Below the graphs, blue and red circles indicate positions of the DSB peaks determined in SET1 dmc1Δ and set1Δ dmc1Δ, respectively. Green dots indicate sites stronger in set1Δ. CYS3 is an example of a DSB site no longer cut in set1Δ, whereas PES4 and SET4 sites are stronger in the absence of SET1.
Dynamics of H3K4 trimethylation during meiosis
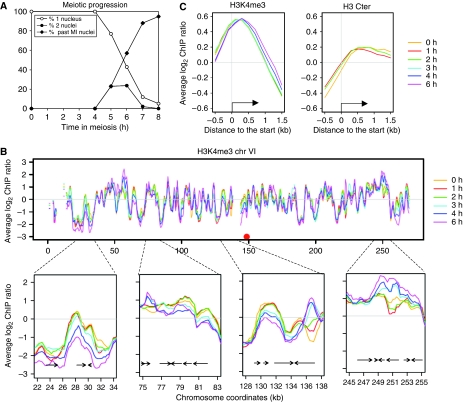
We characterized the levels of H3K4me3 residue as well as total histone H3 associated with chromatin in a S. cerevisiae wild-type diploid strain by ChIP-chip. The synchronous progression in meiosis was verified by fluorescence microscopy of DAPI-stained nuclei (Figure 2A). Western blot analysis showed that amounts of H3K4me3 relative to total histone H3 do not change significantly during the meiotic time course (Supplementary Figure S3). When plotted along chromosomes, the H3K4me3 levels associated with chromatin are relatively constant at most loci during meiosis, but some loci exhibit strong temporal variation (Figure 2B). When plotted as a function of their position relative to the translational start sites of the yeast genes, H3K4me3 is maximal in the first 500 bp of ORFs, and histone H3 is depleted from promoter regions and covers ORFs uniformly (Figure 2C), as is seen in exponentially growing cells (Pokholok et al, 2005). The functional significance of these results is analysed below.
Figure 2.
Distribution of histone H3K4me3 during meiosis. (A) Meiotic progression of the wild-type strain (ORD7339) used for the histone H3 and H3K4me3 ChIP-chip experiments. Meiotic divisions of cells transferred to sporulation medium at t=0 h were monitored by fluorescence microscopy of 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells. (B) H3K4me3 distribution during wild-type meiosis along chromosome VI. Profiles are smoothed by a sliding 1 kb window computed every 250 bp. Four examples of regions where the trimethylation pattern varies are enlarged. (C) Average profiles of H3K4me3 or total histone H3 association with chromatin as a function of position relative to the translational start site. All the genes were aligned according to their start site and probes were grouped in bins of 0.2 kb. Average values for each bin are plotted for each indicated time point.
Refinement of the transcriptional program of meiosis
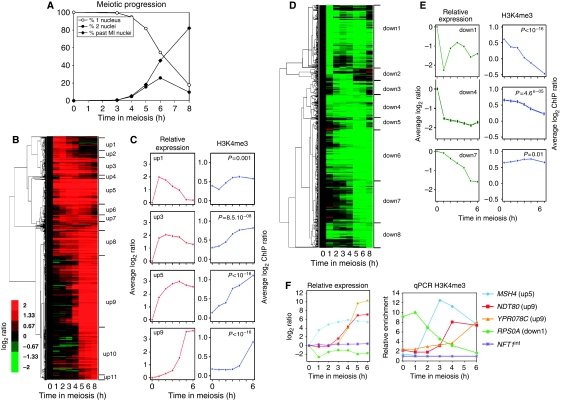
As H3K4me3 is a mark indicative of the transcriptional state of genes during vegetative growth, we first examined the relationships between trimethylation and gene expression, detected by measuring stable mRNA levels, during meiosis. As substantial discrepancies exist between previous studies of meiotic gene expression (Chu et al, 1998; Primig et al, 2000; Friedlander et al, 2006), we independently measured meiotic mRNA level patterns in our wild-type SK1 strain (Figure 3A). We identified 1074 upregulated and 723 downregulated genes, indicating that at least 29% of the genes are transcriptionally regulated during meiosis and sporulation (Figures 3B and D). A total of 85 and 86% of our up- and downregulated genes, respectively, were found in at least one of the previous meiotic regulation studies (see detailed comparisons in Supplementary Figure S4), suggesting the robustness of our list of genes. We classified our set of meiotically regulated genes into groups with similar mRNA level profiles (Supplementary data). Upregulated genes fell into 11 groups (‘up1' to ‘up11'), with a high degree of correlation within each group (Pearson coefficient ⩾0.9) (Figure 3B, Supplementary Table S4). Downregulated genes did not cluster as discretely, but eight groups (‘down1' to ‘down8') were assigned (Figure 3D, Supplementary Table S5).
Figure 3.
Links between meiotic transcriptional gene regulation and histone H3K4 trimethylation. (A) Meiotic progression of the wild-type strain (ORD8622) used for the transcriptome analysis. The values presented are the average of the three independent time courses used to generate the transcriptome data. (B) Hierarchical clustering of the 1074 meiotically upregulated genes. Genes are grouped according to their induction pattern. The name of the clusters up1–up11 is indicated. The colour code reflects the quantitative change of expression relative to time 0 h. (C) Left panels: average expression change relative to time 0 h along meiosis for a subset of upregulated clusters. Right panels: on the 0–500 bp region of each gene of the same clusters, the average level of H3K4 trimethylation was estimated after ChIP and microarray hybridization and represented as a function of time during meiosis. (D) Hierarchical clustering of the 723 downregulated genes. Eight clusters down1–down8 are defined. Other legends as in (B). (E) Average expression changes along meiosis for a subset of downregulated genes clusters. Other legends as in (C). (F) Meiotic variations of H3K4me3 in selected meiotically induced or repressed genes. The relative expression profile obtained from the transcriptome analysis is shown for each gene for comparison (left). Quantitative measurement of H3K4me3 by qPCR (right). Enrichment values were normalized to the enrichment value of NFT1int, an internal sequence of the large NFT1 gene, used for background H3K4me3 control.
Meiotic transcriptional regulation is accompanied by changes in H3K4 trimethylation
To compare the temporal variation of H3K4me3 and transcription, we compared transcriptome and ChIP-chip data from cultures that showed similar kinetics of meiotic progression (Figures 2A and 3A). For each meiotically regulated gene, we calculated the mean H3K4me3 enrichment value observed in the 0–500 bp region of its coding part (Supplementary Table S6). These values were then used to compare the mean temporal mRNA level profiles with the mean trimethylation profile for each transcriptional cluster.
Upregulated genes showed a net increase in H3K4 trimethylation during meiotic progression (Figures 3C and Supplementary Figure S5). A more detailed examination of the data showed that, among upregulated genes, the increase in H3K4me3 levels generally occurred after the increase in transcripts. For genes of cluster up1, which are strongly induced between times 0 and 1 h, an increase in H3K4me3 was observed at 2 h and later (Figure 3C). For most of the other clusters (up2, 3, 5, 6, 7, 8, 9 and 10), we also observed an increase of H3K4me3 that was delayed relative to transcripts (Figure 3C and Supplementary Figure S5). This general pattern was confirmed by qPCR for several individual genes (Figure 3F). For the cluster up11, which contains late meiosis genes, no increase of H3K4me3 was observed. We cannot exclude the possibility that an increase occurs after 6 h, which is the last time point that we have examined. Another exception is the cluster up4, which contains the eight histone coding genes, strongly induced at the time of meiotic replication (2 h) but showing no H3K4me3 increase (Supplementary Figure S5).
Within the clusters down1, 2, 4, 6 and 8 (comprising 73% of the downregulated genes), we noted that the level of H3K4me3 slowly decreases with time (Figures 3E, F and Supplementary Figure S5). This is most evident in cluster down1, in which stable mRNA levels strongly drop during the first hour, but H3K4me3 levels gradually decrease until t=6 h. Taken together, these results show at the genome-wide level that the H3K4 trimethylation is a very dynamic mark, which can be removed from chromatin upon transcription repression. By contrast, some clusters do not show altered H3K4me3 levels during repression (cluster 7, Figure 3E), showing that, in this case, decrease in the amounts of mRNA levels can occur without a substantial alteration in levels of H3K4me3.
H3K4 trimethylation marks meiotic DSB sites, independently of local transcriptional levels
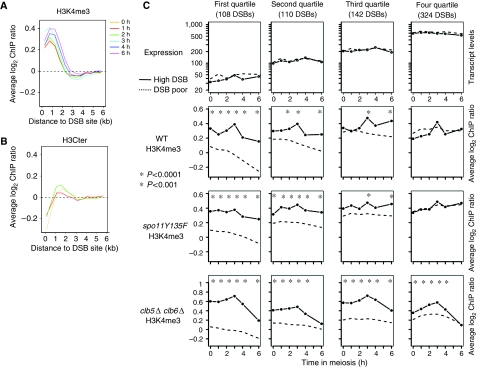
The fact that DSB formation seems to depend on histone H3K4 methylation prompted us to investigate the pattern of H3K4me3 in the proximity of DSB sites. During the whole meiotic time course, H3K4me3 levels are higher close to the DSB sites and decrease with increasing distance from a DSB site (Figure 4A). As H3K4me3 is mainly located on the beginning of the genes, this colocalization could be due to the preferential, if not exclusive, localization of the DSBs in promoter regions (Baudat and Nicolas, 1997; Gerton et al, 2000; Blitzblau et al, 2007). This is exemplified by the fact that total H3 levels are low close to DSB sites compared with the rest of the genome (Figure 4B). To exclude a potential influence of transcription on H3K4 trimethylation variation close to DSB sites, we analysed the data as follows. First, we examined H3K4me3 levels in regions located near the 4361 genes that are neither up- or downregulated but exhibit a constant level of transcripts during meiosis (non-regulated), representing 71% of the genes. Second, to distinguish between transcription-dependent and transcription-independent levels of H3K4me3 close to DSB sites, we compared the set of the 1013 hottest DSB regions (DSB-high) with a similar set of control regions that did not display any of these hot spot DSBs (DSB-poor; Supplementary data). To avoid a strong influence of high transcription on H3K4me3 close to DSB sites, we subclassified the non-regulated genes into four quartiles, according to their average absolute transcript levels during meiosis. For this, we used another set of transcriptome data, the one from the study of Primig et al (2000), because they obtained not relative, but absolute quantitative data of transcript levels during meiosis. Within each quartile, similar transcript levels were seen for genes that are close to a DSB hot spot and genes that are not (Figure 4C, upper panels). In contrast, the level of H3K4me3 close to the DSB hot spots is very similar and ‘constitutively' high in all quartiles, whereas in the DSB-poor regions, the average level of H3K4me3 showed a strong dependence on transcript levels and was lower than in DSB-high regions (except in the most highly transcribed quartile). The same conclusion was reached when H3K4me3 levels were normalized to total H3 levels (Supplementary Figure S6). Thus, in addition to being a mark of transcription, H3K4me3 also marks meiotic recombination initiation regions.
Figure 4.
DSB sites are constitutively hypertrimethylated, independently of transcript levels. (A) Average H3K4me3 as a function of the distance from the DSB sites. All the 1013 DSB hot spot sites were aligned and probes were grouped in bins of 0.5 kb. Average values for each bin are plotted for each indicated time point. (B) Average total H3 levels as a function of the distance from the DSB sites. Same legend as in (A). (C) The non-regulated genes were divided into four quartiles according to their average transcript levels during meiosis. Then DSB sites were examined in each category, both for expression levels of the adjacent genes (top panel), and for H3K4me3 (bottom panels). Profiles of control DSB-poor regions were also calculated for each quartile (dotted lines), as described in Supplementary data. Stars indicate a significant difference between DSB-high and DSB-poor regions. WT, ORD7339; spo11Y135F, ORD7341; clb5Δ clb6Δ, ORD6830.
Elevated H3K4me3 levels near DSB sites are independent of replication and DSB formation
To ask if the high level of H3K4me3 close to DSB sites was influenced by DSB formation, we examined the meiotic H3K4me3 profiles and transcriptomes in the spo11Y135F mutant, which is deficient in meiotic DSB formation (Bergerat et al, 1997) and progresses through meiosis to form inviable spores (Supplementary Figure S7). Patterns of meiotic transcription in wild type and spo11Y135F strains are globally similar (Supplementary Table S6 and Figure S7) and similar H3K4me3 profiles are observed in the two strains, with few exceptions, such as cluster down5 (Supplementary Figure S5). Finally, the average level of H3K4me3 is greater near DSB-high regions, independently of the absolute level of gene expression, as in wild-type cells (Figure 4C). Next, we investigated the effect of premeiotic replication by using the clb5Δ clb6Δ mutant, deficient in the premeiotic replication that normally precedes DSB formation by 2 h (Stuart and Wittenberg, 1998), and which shows no meiotic DSB formation (Smith et al, 2001). In these cells, the expression of the early genes of clusters up1–8 is not affected, and the gene clusters that are induced later in meiosis (up9–11) show reduced expression, consistent with the impairment of cell progression past MI (Supplementary Figure S7 and Table S6). H3K4me3 levels in the clb5Δ clb6Δ cells, as in WT, are greater near DSB-high regions in all expression quartiles (Figure 4C). We conclude that the H3K4me3 mark occurs preferentially near DSB sites in a manner that is independent of DSB formation and meiotic DNA replication.
H3K4me3 marks natural DSB regions before entry into meiosis
As meiotic replication, the major chromosomal event that precedes DSB formation, does not set the H3K4me3 difference between DSB-high and DSB-poor regions, we asked when this differential mark is established. To determine whether it pre-exists before entry into meiosis, we examined the H3K4me3 profile of a wild-type strain growing exponentially in rich medium (YPD). Remarkably, as in meiotic cells, H3K4me3 was enriched in DSB-high regions in each of the transcription quartiles, whereas in DSB-poor regions, the H3K4me3 levels co-varied with transcript levels (Figure 5, top panel). A higher H3K4me3 level was also seen for DSB sites showing a lower enrichment, between two- and five-fold over background (DSB-mild, Supplementary Figure S8). We excluded the possibility that the difference between DSB-high and DSB-poor regions was due to differences in transcript levels measured in exponentially growing cells (Holstege et al, 1998); as in meiosis, the future DSB regions are not located in the vicinity of more transcribed genes than control DSB-poor regions (Figure 5, lower panel). Therefore, H3K4me3 is a differential mark of potential meiotic recombination initiation sites that exists before the induction of meiosis.
Figure 5.
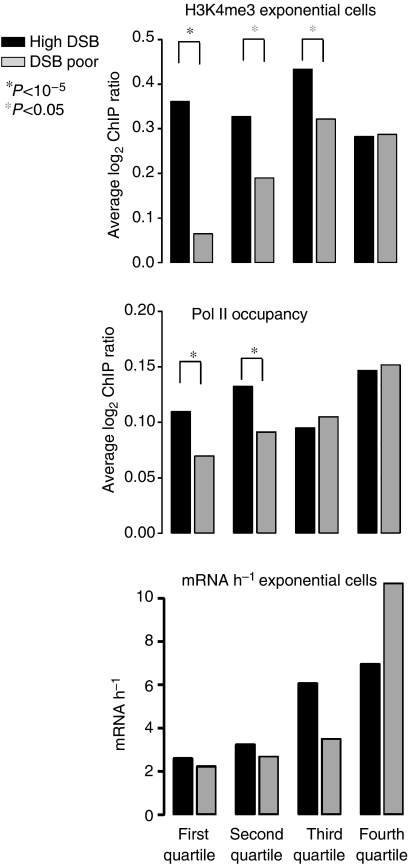
Hypertrimethylation of future DSB sites preexists in exponentially growing cells, and shows more RNA Polymerase II occupancy. For each of the same meiotic transcription quartiles as in Figure 4, we profiled H3K4me3 in exponentially WT growing cells (ORD7339, top panel). In the middle panel, we determined Pol II occupancy, according to published results (Steinmetz et al, 2006), close or not to DSB sites, in each quartile. The bottom panel shows, in each quartile, the transcription levels determined in exponentially growing cells (Holstege et al, 1998), close or not to a DSB site.
Higher RNA polymerase II occupancy in DSB regions
As high levels of H3K4me3 pre-exist in DSB regions located near low mRNA abundance genes (first quartile, Figure 4C, top panel), we wondered whether the H3K4me3 mark might be nevertheless deposited by RNA polymerase II, but without giving rise to detectable coding transcripts. We therefore compared the Pol II occupancy (Steinmetz et al, 2006) in our sets of DSB and DSB-poor regions as a function of transcription quartile. Intriguingly, in the two lowest mRNA-abundance quartiles, we found a highly significant (both P<10−5) increase in Pol II occupancy in DSB regions than in equivalent DSB-poor regions (Figure 5, middle panel). This may be the source of chromatin events leaving the H3K4me3 mark (see Discussion).
H3K4me3 preferentially marks regions where DSB can be targeted
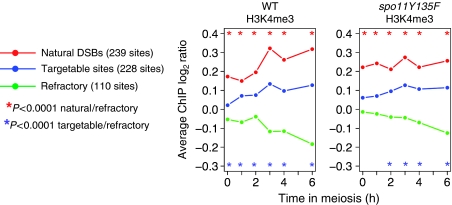
The formation of meiotic DSBs in some normally DSB-poor chromosomal regions can be stimulated by the forced recruitment of a Gal4BD–Spo11 fusion protein to naturally occurring Gal4-binding sequences. These regions are defined as ‘targetable'. In contrast, in other regions, the binding of Gal4BD–Spo11 is not sufficient to trigger DSB formation, and these regions are called ‘refractory' (Robine et al, 2007). QQR–Spo11 is another fusion of Spo11 to three artificial zinc fingers targeting DSBs in other regions than Gal4BD–Spo11. We compared the levels of H3K4me3 in the set of natural, targetable and refractory DSB regions, identified in ChIP-chip analyses of DSB formation in wild-type, Gal4BD–Spo11 and QQR–Spo11 cells (Robine et al, 2007, Uematsu et al, unpublished data). Both in the wild-type and spo11Y135F strains, the natural DSB regions exhibit the highest level of H3K4me3, the targetable regions are intermediate and the refractory regions show the lowest level of H3K4me3 (Figure 6). Thus, the potential of the DSB-poor regions to be targeted (provided that a functional Spo11 protein is brought there), similar to naturally occurring DSB regions, is also correlated with the levels of H3K4me3.
Figure 6.
H3K4 trimethylation levels close to the natural DSB or targetable sites. Average H3K4me3 levels are compared among regions where a DSB made by the endogenous Spo11 is observed (natural), those where DSB can be artificially introduced when Gal4BD–Spo11 or QQR–Spo11 is targeted there (targetable) and those where Gal4BD–Spo11 or QQR–Spo11 is targeted but does not cleave (refractory). The different categories of regions were defined by Robine et al (2007) and Uematsu et al (unpublished data). To obtain the H3K4me3 profile of each category along meiosis, we averaged the H3K4me3 level of all the probes spanning the defined DSB regions in each category. Stars indicate a significant difference between the indicated categories of regions (Wilcoxon test).
The absence of H3K4 trimethylation most strongly impairs formation of DSB at sites that were highly trimethylated
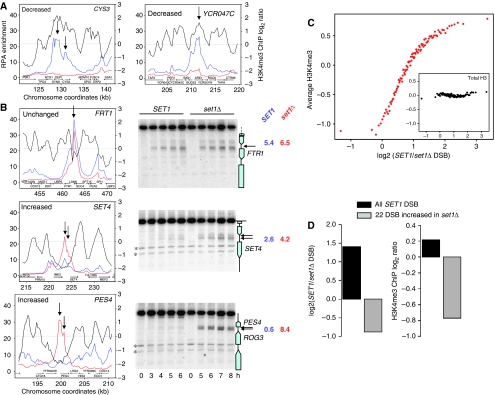
Although the absence of Set1 decreases DSB frequencies in at least 84% of the natural sites, not all DSB sites are affected to the same extent. To investigate the reasons for these differences, we examined the wild-type H3K4me3 levels in three types of DSB regions: strongly reduced in set1Δ (CYS3 and YCR047C; Figure 7A), not affected by set1Δ (FTR1; Figure 7B) and increased in set1Δ (SET4 and PES4; Figure 7B). In all these cases, Southern blot analyses confirmed the variations of DSB frequencies in wild-type versus set1Δ strains identified by CHIP-chip and showed that the DSBs increased in set1Δ are located in an intergenic region, similar to DSBs occurring in wild-type cells (Figure 7B). Notably, at the set1Δ-specific PES4 locus, DSB frequency in the set1Δ cell (8.4%) reaches the level of the strongest DSB sites in wild-type cells (e.g., at YCR047C, Figure 1A). As expected, the strongly reduced DSB sites coincide with high H3K4me3 peaks (Figure 7A), but strikingly the two sites with increased DSBs in set1Δ localize in an H3K4me3 ‘valley' and the unchanged DSB occurs at a site where H3K4me3 level was intermediate (Figure 7B, right panels). To extend these indications to the entire genome, we arranged all the array elements in groups with increasing variation of DSB signal in set1Δ and looked for each group at the H3K4me3 levels measured in exponentially growing wild-type cells (Figure 7C). There is a very strong correlation between H3K4me3 levels and the way DSB formation will be affected by set1Δ: very highly trimethylated sites have a strongly reduced DSB signal in set1Δ, the few sites that were occurring in low trimethylated regions are less affected, and even more, some new sites appear in trimethylation ‘deserts'. Importantly, total H3 levels are similar whether or not the region is affected by set1Δ for DSB formation (Figure 7C, inset), indicating the specific function of H3K4 trimethylation in specifying the fate of DSB formation.
Figure 7.
Correlation between DSB variation in set1Δ versus wild-type cells and local H3K4me3 levels. (A) DSB formation at the YCR047C (BUD23) and the CYS3 loci occurs in an H3K4me3 peak and is strongly reduced in the absence of Set1. Left y axis: DSB enrichment for SET1 dmc1Δ (blue) and set1Δ dmc1Δ (red). Right y axis: H3K4me3 enrichment from exponentially growing WT cells (ORD7339). The arrow shows the DSB site. (B) Example of DSB sites not affected by set1Δ, in the FTR1 promoter, or increased in set1Δ (SET4 and PES4). Left: graph showing the properties of the locus, as in (A). Right: Southern blot measurement of DSB formation at the locus, both in SET1 and in set1Δ. Restriction digest and probes are in Supplementary data. In both panels, the arrows show the DSB sites. On the right of each blot is indicated the maximal DSB frequency observed in each strain. (C) All the significant 41500 array elements were grouped in 100 groups of 415 probes according to their ratio of dmc1Δ/set1Δ dmc1Δ RPA enrichment. Each dot represents one such group. For each group, arranged according to this ratio (x axis), the average H3K4me3 level measured in exponentially growing cells is indicated (y axis, top panel). Inset: average levels of total histone H3 measured at t=0 h of meiosis in the same groups, showing that the effect observed is linked specifically to H3K4 trimethylation and not to varying histone H3 levels. (D) Characteristics of the 22 sites that show increased DSB frequency in set1Δ, in terms of increase in DSB frequency compared with SET1 (left panel) and H3K4me3 levels, measured in exponentially growing cells (right panel).
Finally, we searched for common characteristics of the 22 sites that have a greater than 1.4-fold increase of DSB signal in set1Δ (listed in Supplementary Table S7). Compared with wild-type DSB sites, we found no difference in terms of proximity to a replication origin, size of the intergenic region and proximity to a meiotically upregulated gene. Their only common feature is to be located in regions of extremely low H3K4me3 (Figure 7D). In summary, in the absence of H3K4me3, few new DSB sites appear, preferentially in regions that are poorly trimethylated in wild-type cells.
Discussion
Here, we report the genome-wide dynamics of H3K4me3 during meiosis. We show that the H3K4me3 levels are highly dynamic, and affected both by transcription induction and transcription repression. Furthermore, once contributions from transcription are filtered, we uncovered a strikingly higher level of H3K4me3 in the preferential regions of recombination initiation. Together with the fact that in the set1Δ cells, totally deficient in H3K4 methylation, the vast majority of DSBs are strongly reduced, this demonstrates the prominent function of this histone modification in the essential and evolutionary conserved process of meiosis.
Temporal and functional relationships between transcription and H3K4 trimethylation
As expected from results in vegetative cells (Morillon et al, 2005), H3K4me3 is increasing in the 5′-part of genes that are transcriptionally activated during meiosis. Our study is the first genome-wide study of the dynamics of H3K4 methylation coupled to transcription analysis. Contrary to what had been seen at a single locus for the MET16 gene, where H3K4me3 appeared very shortly after MET16 activation (Morillon et al, 2005), we clearly detect an increase of transcripts before we can detect an increase of H3K4 trimethylation. Thus, H3K4me3 levels seem to follow meiotic transcriptional activation, in agreement with the fact that, in set1Δ cells, the absence of H3K4 methylation does not have a strong effect on gene induction during meiosis (Sollier et al, 2004). As recently observed in studies of human cells (Sims et al, 2007), the H3K4 trimethylation mark in yeast may be involved in maintaining the activated state or in post-initiation processes.
Another observation is that H3K4me3 levels decrease in some of the gene clusters where transcript levels drop rapidly during meiosis (i.e., down1, down4 and down6), and a similar decrease in H3K4 trimethylation is seen in meiotic replication-deficient clb5Δ clb6Δ cells (Supplementary Figure S5). Thus, the trimethylation loss is not due to replacement by unmethylated histones during replication. The loss of trimethylation must therefore occur either through active demethylation or by replication-independent histone turnover. Conversely, other clusters showing a drop of transcript levels maintain high levels of H3K4me3. This may reflect another type of regulation occurring by continued transcription, and thus continuous H3K4me3 deposition, but increased mRNA turnover. Alternatively, transcription might be blocked but H3K4me3 not removed from these genes. In any case, keeping high levels of H3K4me3 in these genes may have an important physiological and adaptative function, as S. cerevisiae cells can undergo meiotic reversal (return to mitotic growth) upon nutritional shift (Zenvirth et al, 1997). In this circumstance, it will be advantageous that the downregulated genes are maintained in a nucleosomal state allowing a rapid reactivation of transcription either from reinitiation or elongation of paused transcripts.
Function of H3K4 trimethylation in the regulation of meiotic DSB formation
Previous studies indicated that an open chromatin configuration is necessary but not sufficient for DSB formation (Wu and Lichten, 1994). Consistently, we observed that H3 occupancy is low in DSB regions and that the level of H3K4me3 is constitutively higher in the vicinity of DSB sites, as compared with the DSB-poor sites.
How does this newly uncovered chromatin factor enrich our understanding of the genome-wide and local controls of DSB formation? The important reduction of DSB frequencies at the vast majority of sites in set1Δ shows that H3K4me3 modification is a common factor involved in DSB formation, irrespective of transcription levels. In contrast, the other studied histone modifications, assayed upon inactivation of the corresponding modifying enzymes, seem to exhibit weaker and more regional effects. Compared with the wild-type strain, the absence of the Sir2 histone deacetylase affects approximately 12% of the DSB sites (5% being elevated and 7% decreased), and the majority of the variations occur close to the ends of the chromosomes (Mieczkowski et al, 2007). Differently, the removal of the Set2 histone methylase- or the Rpd3 histone deacetylase-modifying enzymes were found to increase DSB frequency at the HIS4 hot spot without any effect on the activity of another hot spot, ARG4 (Merker et al, 2008). Thus, the effect of these histone modifications is most likely dependent on the particularity of each region, whereas the H3K4 methylation has a ubiquitous function. The function of H3K4me3 in DSB formation might be indirect by regulating the accessibility of the chromatin substrate to Spo11 and the other DSB-forming factors, either by recruiting chromatin remodellers, such as the NURF complex (Li et al, 2006; Martin et al, 2006) or by antagonizing Sir-dependent silencing (Venkatasubrahmanyam et al, 2007). Alternatively, as for V(D)J recombination (Liu et al, 2007; Matthews et al, 2007), the function of the H3K4me3 mark can be direct in recruiting one or several of the factors involved in DSB formation.
The functional importance of the level of H3K4me3 in controlling DSB formation also bears from several other observations reported here: in particular, the fact that it is not dependent on meiotic replication and pre-exists in vegetative cells. Thus, with the light of the recent findings that DSB formation is controlled by post-translational phosphorylation of the Mer2 DSB protein (reviewed in Murakami and Keeney, 2008), DSB formation seems to depend on two parallel processes: one setting before meiosis this and perhaps other chromosomal determinants required for DSB formation, whereas the second layer of regulation assembles and activates the DSB machinery according to the progression of meiosis and monitoring the meiotic replication to avoid the formation of DSB in non-replicated substrates.
DSB formation in the absence of Set1
We find that the only common feature shared by the 22 sites with stronger DSB formation in the absence of H3K4me3 is a very low level of this histone mark in wild-type cells. As previous studies showed that DSB formation at a given site is sensitive to long-distance cis-acting controls (e.g., Robine et al, 2007), a simple explanation would be that the inhibitory effect of the neighbouring DSB sites is removed in the absence of Set1 because of their reduced DSB frequencies. Another non-exclusive hypothesis is that very low endogenous H3K4me3 levels allow for the binding of other chromatin factors, which are also able to attract the DSB-forming machinery. It will be interesting to ask whether or not meiotic DSB formation at these Set1-independent sites still depends on the other gene products normally required for DSB formation in wild-type cells.
Source of the H3K4 hypertrimethylation of potential DSB sites
H3K4me3 levels in DSB-forming regions are high and similar in weakly and highly transcribed regions, and these high levels already exist in exponentially growing wild-type cells. The source of this H3K4 trimethylation, which is not meiosis-specific, is puzzling. One possibility is that the H3K4 trimethylation at future DSB sites is not formed by the transcription machinery, but by other unknown chromatin-level activities. Alternatively, it may reflect the production of unstable transcripts that would be underrepresented in analyses of mRNA steady-state levels. Recent studies have shown that in vegetative cells, RNA polymerase II is present in the vast majority of the genome, including many regions considered to be transcriptionally inert (Steinmetz et al, 2006), and that many Pol II-dependent RNAs are degraded by members of the nuclear exosome and the TRAMP complex (LaCava et al, 2005; Wyers et al, 2005). As the Set1 methylase travels with elongating Pol II (Ng et al, 2003), these regions would be expected to be methylated, even though the resulting mRNAs would not be detected. The high level of H3K4me3 and the significantly higher Pol II occupancy (Figure 5) may occur close to poorly transcribed genes because of poised RNA polymerase emanating from cryptic promoters. Having now identified a large subset of DSB sites with no detectable transcripts, novel analyses can be conducted to ultimately describe each layer of DSB control.
New avenues for understanding the choice of DSB sites
Genome-wide mapping of the Spo11-dependent DSBs clearly indicates that their distribution is not random along the chromosomes (Figures 1 and Supplementary Figure S2). Our results indicating that pre-conditions for DSB formation rely on histone modifications and/or transcriptional activity occurring during the preceding vegetative growth raise the interesting possibility that it is modulated by the environmental growth conditions. To date, a limited number of studies have addressed such issues. Genome-wide mapping of DSBs revealed a significant overrepresentation of DSB hot spots at genes in the metabolism-functional class, particularly at genes involving amino-acid biosynthesis (Gerton et al, 2000). In addition, external conditions have been reported to affect recombination rates at certain loci (Koren et al, 2002). For example, meiotic recombination hot spot activity at HIS4 is modulated by the binding of transcription factors (Petes, 2001) and auxotrophy for lysine or adenine (Abdullah and Borts, 2001), and it shows a strong dependence on temperature, increasing when the temperature is shifted to 18°C (Nag et al, 1989). The inherent flexibility of such physiological control, acting at the level of chromatin, is well suited to explain two unresolved features of meiotic recombination initiation in yeast and other organisms. First, the fact that DSBs occur stochastically along the chromosomes may simply translate differences in the metabolic state of individual cells. Second, recombination hot spots may have evolved rapidly in a manner disproportionate to the change of DNA sequence, as recently found between chimpanzees and humans (Winckler et al, 2005), because the regulation at the chromatin level is a prominent control factor and is more malleable than the local change of DNA sequence.
Materials and methods
Yeast strains and culture conditions
Genotypes of the SK1 S. cerevisiae strains used here and culture conditions are provided in Supplementary data.
mRNA isolation, cDNA target preparation and microarray hybridization
mRNA was isolated from 40 ml of culture as described in Supplementary data. A quantity of 2 μg of mRNA from each time point was used for cDNA synthesis with incorporation of aminoallyl-dUTP (Sigma), then labelled by coupling to the Cy5 dye (Amersham) as described at http://cat.ucsf.edu/resources/index.html. A reference mRNA mix was prepared for each kinetics experiment by pooling equal amounts of mRNA from every time-point of the time course, except for t=0 h, which was added at four times the amount of the other samples. cDNA was prepared from the pooled reference mix, labelled with Cy3 dye, aliquoted and hybridized against the Cy5-labelled probes from each time point. Hybridization to yeast ORFs microarrays and washes were performed as described previously (Borde et al, 2004).
Chromatin immunoprecipitation and microarray hybridization
A total of 20 ml (4 × 108 cells) were crosslinked with formaldehyde and chromatin immunoprecipitation was performed as described (Robine et al, 2007), using 3 μl of rabbit polyclonal anti-H3K4me3 (Abcam no. Ab8580), 3 μl of rabbit polyclonal anti H3Cter (Abcam no. 1791) or 2.5 μl of a rabbit polyclonal anti-Rfa1 antibody (given by Steven Brill) and 30 μl protein G magnetic beads suspension (Dynal). For microarray hybridizations, two-thirds of the immunoprecipitated DNA or 1/70 of the DNA from the whole-cell extract was amplified as described (Robine et al, 2007). A total of 1 μg of each sample was coupled with either Cy3 (whole-cell extract) or Cy5 (immunoprecipitated sample) and hybridized on an Agilent 44k yeast whole genome oligonucleotide array, which contains probes spaced on average every 290 bp (Agilent, no. G4493A) for 16 h at 65°C in the 1 × hybridization buffer supplied by Agilent. Slides were washed as described by Buhler et al (2007). ChIP quantification by qPCR is described in Supplementary data and primers are listed in Supplementary Table S8.
Data acquisition and analysis
Microarray images were acquired using a GenePix 4000 scanner and data were quantified using GenePix Pro 4.0 or 5.1 software (Axon Instruments). For all microarray experiments, the median local background intensity was subtracted from the median spot intensity before calculating for each spot the intensity ratio (Cy5/Cy3 median intensity). For transcriptome analyses, these ratios, representing the transcript abundance relative to the reference mix, were normalized using the Lowess algorithm implemented in BASE (BioArray Software Environment) 1.0.7 program (Saal et al, 2002; Yang et al, 2002). For each time point of a time course, we zero-centered the data by calculating the expression ratio relative to the t=0 h transcription for further analysis and comparison with other data. Data were missing at t=6 h in one of the wild-type time courses. As a substitute, values were interpolated linearly from the data at t=5 h and t=8 h. Methods for clustering the gene expression profiles are described in the legend of Supplementary Figure S6. For ChIP-chip of H3K4me3, total H3 and RPA analyses, we used different normalization methods described in Supplementary data.
Calculation of gene expression levels
For this purpose, we used the data of Primig et al (2000). We attributed to each of the 4844 non-regulated genes the mean absolute expression value measured all along the time course (from t=0 h to t=10 h). Then, the 4844 genes were divided into four quartiles (each with 1211 genes) depending on their mean expression value. Next, each DSB site was assigned a neighbour with a mean expression value: when located in an ORF, a DSB site was associated with the mean expression value of the corresponding gene. When located in a promoter, it was associated with the mean expression value of the downstream gene, and when located in the promoters of two divergent genes, the DSB was assigned the maximum of the mean expression value of the two divergent genes. Thus, by construction, there are more DSBs in the most highly transcribed quartile than in the least one.
Definition of the DSB-poor reference regions
For ‘DSB sites', we calculated the mean H3K4me3 or H3total ratio of all probes located less than 300 bp around the DSB position. For the reference (DSB-poor sites) regions, we picked random sets of the same number of probes as the ones for DSB sites, not close to a DSB, but with the same distribution with respect to a start codon as the set of probes close to DSB sites. Finally, we computed the average of H3K4me3 or H3total enrichment values for 100 of these random sets, and used it as the non-DSB ratio. Thus, the only difference between the two populations is the proximity to a DSB. To test the significance of the observed differences, we calculated the Wilcoxon signed-rank test for each of the 100 simulations and corrected for multiple tests.
Microarray data
The raw data of all our microarray experiments are available at the GEO database website, accession number GSE11004.
Supplementary Material
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Supplementary Table 7
Supplementary Table 8
Supplementary Tables 9
Supplementary Figures and Information
Acknowledgments
We thank Nelly Balège for performing the spo11Y135F transcriptome experiment and Steven Brill for anti-Rfa1 antibody. We thank Michael Lichten for helpful comments. This work was supported by grants from the French Ministry of Research and Technology ACI no. BCM0098 program to VG and AN, the ANR (BLANC06-3-150811) to AN, Ligue Nationale contre le Cancer to VG and AN and the INSERM (PNRRE ‘Jeunes Chercheurs') to VB. NR and WL were supported by pre-doctoral fellowships from the ARC.
References
- Abdullah MF, Borts RH (2001) Meiotic recombination frequencies are affected by nutritional states in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 98: 14524–14529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Baudat F, Nicolas A (1997) Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci USA 94: 5213–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA 99: 8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, Schreiber SL, Lander ES (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A (2007) Mapping of meiotic single-stranded DNA reveals double-strand-break hotspots near centromeres and telomeres. Curr Biol 17: 2003–2012 [DOI] [PubMed] [Google Scholar]
- Borde V, Lin W, Novikov E, Petrini JH, Lichten M, Nicolas A (2004) Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell 13: 389–401 [DOI] [PubMed] [Google Scholar]
- Buhler C, Borde V, Lichten M (2007) Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I (1998) The transcriptional program of sporulation in budding yeast. Science 282: 699–705 [DOI] [PubMed] [Google Scholar]
- Dehe PM, Geli V (2006) The multiple faces of Set1. Biochem Cell Biol 84: 536–548 [DOI] [PubMed] [Google Scholar]
- Friedlander G, Joseph-Strauss D, Carmi M, Zenvirth D, Simchen G, Barkai N (2006) Modulation of the transcription regulatory program in yeast cells committed to sporulation. Genome Biol 7: R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD (2000) Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA 97: 11383–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hall H, Hunt P (2007) The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet 16 (Spec No. 2): R203–R208 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Johnson R, Borde V, Neale MJ, Bishop-Bailey A, North M, Harris S, Nicolas A, Goldman AS (2007) Excess single-stranded DNA inhibits meiotic double-strand break repair. PLoS Genet 3: e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Ben-Aroya S, Kupiec M (2002) Control of meiotic recombination initiation: a role for the environment? Curr Genet 42: 129–139 [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ (2006) Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442: 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S (2007) A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity 27: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L (2006) The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol 26: 7871–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA (2007) RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450: 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker JD, Dominska M, Greenwell PW, Rinella E, Bouck DC, Shibata Y, Strahl BD, Mieczkowski P, Petes TD (2008) The histone methylase Set2p and the histone deacetylase Rpd3p repress meiotic recombination at the HIS4 meiotic recombination hotspot in Saccharomyces cerevisiae. DNA Repair (Amst) 7: 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Dominska M, Buck MJ, Lieb JD, Petes TD (2007) Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104: 3955–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J (2005) Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell 18: 723–734 [DOI] [PubMed] [Google Scholar]
- Murakami H, Borde V, Shibata T, Lichten M, Ohta K (2003) Correlation between premeiotic DNA replication and chromatin transition at yeast recombination initiation sites. Nucleic Acids Res 31: 4085–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Keeney S (2008) Regulating the formation of DNA double-strand breaks in meiosis. Genes Dev 22: 286–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag DK, White MA, Petes TD (1989) Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature 340: 318–320 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Ohta K, Shibata T, Nicolas A (1994) Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J 13: 5754–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2: 360–369 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE (2000) The core meiotic transcriptome in budding yeasts. Nat Genet 26: 415–423 [DOI] [PubMed] [Google Scholar]
- Reddy KC, Villeneuve AM (2004) C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452 [DOI] [PubMed] [Google Scholar]
- Robine N, Uematsu N, Amiot F, Gidrol X, Barillot E, Nicolas A, Borde V (2007) Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 27: 1868–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Saal LH, Troein C, Vallon-Christersson J, Gruvberger S, Borg A, Peterson C (2002) BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol 3: SOFTWARE0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2004) Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 6: 73–77 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D (2007) Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Penkner A, Ohta K, Klein F, Nicolas A (2001) B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr Biol 11: 88–97 [DOI] [PubMed] [Google Scholar]
- Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Geli V, de La Roche Saint-Andre C (2004) Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J 23: 1957–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA (2006) Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell 24: 735–746 [DOI] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C (1998) CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev 12: 2698–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD (2007) Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z.. Proc Natl Acad Sci USA 104: 16609–16614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler W, Myers SR, Richter DJ, Onofrio RC, McDonald GJ, Bontrop RE, McVean GA, Gabriel SB, Reich D, Donnelly P, Altshuler D (2005) Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308: 107–111 [DOI] [PubMed] [Google Scholar]
- Wu TC, Lichten M (1994) Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518 [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A (2005) Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K (2004) Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J 23: 1792–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Shinohara M, Shinohara A (2004) Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci USA 101: 11380–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth D, Loidl J, Klein S, Arbel A, Shemesh R, Simchen G (1997) Switching yeast from meiosis to mitosis: double-strand break repair, recombination and synaptonemal complex. Genes Cells 2: 487–498 [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Supplementary Table 7
Supplementary Table 8
Supplementary Tables 9
Supplementary Figures and Information