Abstract
LmrR is a PadR-related transcriptional repressor that regulates the production of LmrCD, a major multidrug ABC transporter in Lactococcus lactis. Transcriptional regulation is presumed to follow a drug-sensitive induction mechanism involving the direct binding of transporter ligands to LmrR. Here, we present crystal structures of LmrR in an apo state and in two drug-bound states complexed with Hoechst 33342 and daunomycin. LmrR shows a common topology containing a typical β-winged helix-turn-helix domain with an additional C-terminal helix involved in dimerization. Its dimeric organization is highly unusual with a flat-shaped hydrophobic pore at the dimer centre serving as a multidrug-binding site. The drugs bind in a similar manner with their aromatic rings sandwiched in between the indole groups of two dimer-related tryptophan residues. Multidrug recognition is facilitated by conformational plasticity and the absence of drug-specific hydrogen bonds. Combined analyses using site-directed mutagenesis, fluorescence-based drug binding and protein–DNA gel shift assays reveal an allosteric coupling between the multidrug- and DNA-binding sites of LmrR that most likely has a function in the induction mechanism.
Keywords: bacterial multidrug transcription factors, multidrug recognition, LmrR, PadR, protein structure
Introduction
Multidrug resistance (MDR) is frequently caused by the action of specialized membrane-bound pumps that possess or have acquired the ability to extrude a wide variety of chemically and structurally different compounds from the cell (Saier et al, 1998; Higgins, 2007). The molecular mechanisms of substrate recognition by these multidrug transporters are poorly understood, mainly because the proteins involved are very recalcitrant towards crystallization, a prerequisite for a detailed structural analysis by X-ray crystallography. Instead, general features explaining multidrug-binding specificity have been derived from structural studies of the transcriptional regulators of multidrug transporters, which are soluble proteins and often bind many of the same diverse drugs that are substrates of the pumps (Zheleznova et al, 1999; Heldwein and Brennan, 2001; Schumacher et al, 2001; Alguel et al, 2007; Higgins, 2007).
LmrR is a recently identified transcription factor that controls the expression of the heterodimeric ABC transporter LmrCD, which is a major multidrug transporter in Lactococcus lactis (Lubelski et al, 2006). It is encoded in the immediate vicinity of the lmrCD genes and was shown to specifically bind to the lmrCD and lmrR promotors where it functions as a transcriptional repressor and autoregulator, respectively (Agustiandari et al, 2008). Toxic compounds that form substrates of the LmrCD transporter include the DNA-binding drugs Hoechst 33342 (H33342), daunomycin, ethidium bromide and rhodamine 6G. One of these drugs, H33342, was shown to directly interact with LmrR, and its presence in the growth medium induced a significant upregulation of the lmrCD genes (Agustiandari et al, 2008). This strongly suggests that the transcription factor may function as a drug sensor causing stimulation of LmrCD production in the presence of toxic compounds, thus promoting their extrusion from the cell. By homology, LmrR belongs to the PadR family of transcriptional regulators found in bacteria and archaea (Gury et al, 2004; Huillet et al, 2006). Only a few PadR family members have been functionally characterized, showing that these proteins have important functions in the regulation of distinct cellular pathways leading, for example, to MDR, virulence and detoxification. Crystal structures of two PadR-like proteins, AphA (De Silva et al, 2005) and Pex (Arita et al, 2007), revealed the characteristics of the PadR fold, which includes a conserved N-terminal winged helix-turn-helix (wHTH) DNA-binding domain (Aravind et al, 2005) that is architecturally similar to that of the multiple antibiotic resistance repressor MarR (Alekshun et al, 2001), and a highly diverse C-terminal helical domain which serves as a dimerization module.
No structural data are available on multidrug binding transcriptional regulators of the PadR family. To understand the structural basis for multidrug recognition by LmrR, as well as to provide insights in the molecular mechanisms involved into the regulation of lmrCD expression, we have determined the crystal structure of LmrR in an apo state, as well as in two drug-bound states, complexed with H33342 and daunomycin.
Results
Structure determination of LmrR in apo and drug-bound states
Full-length LmrR was purified both as a fusion protein containing a C-terminal streptactin tag and as untagged protein. The crystal structure of untagged LmrR was determined at 2.0-Å resolution, by the molecular replacement method using an ensemble of three structurally homologous, but functionally uncharacterized proteins from the Protein Data Bank (PDB) as a search model. The untagged LmrR structure was solved in the absence of bound drugs and thus represents an apo form. Tagged LmrR was also crystallized in the absence of drugs, and its structure was determined at 2.5-Å resolution. As the overall structural features of drug-free tagged LmrR are identical to that of drug-free untagged LmrR, we will describe the latter structure only and refer to it as apo–LmrR.
Crystals of drug-bound LmrR were obtained by co-crystallization using tagged and untagged LmrR preincubated with different lipophilic cationic drugs. Co-crystals of tagged LmrR complexed with H33342 and with daunomycin diffracted both to 2.2-Å resolution. Co-crystals obtained with untagged LmrR, or in the presence of ethidium bromide and rhodamine 6G, could not be used for structure determination because of their poor X-ray diffraction quality. In all cases, however, the presence of bound drug was indicated by a drug-specific colouring of the crystals. The structures of LmrR bound to H33342 and daunomycin were determined by molecular replacement using the apo–LmrR subunit structure as a search model.
Each of the three LmrR structures (apo, H33342-bound and daunomycin-bound) represents a different crystal form with one or two LmrR subunits in the asymmetric unit. In all crystals, LmrR is present as dimers formed through either crystallographic or non-crystallographic symmetry. The dimeric nature of LmrR is consistent with the results of gel filtration chromatography and dynamic light scattering experiments (not shown), and agrees with the general oligomeric preference of other MarR/PadR family members. The overall fold of the LmrR dimer is the same as in the different structures, but structural superpositions reveal some notable differences, highlighting a significant inherent conformational plasticity (further explained below). The stereochemical quality of the models is excellent with no Ramachandran outliers. The electron density for the polypeptide chains is generally well defined, except for the N and C termini, including the streptactin tag, which are disordered and not included in the final models. Another flexible region that is disordered in most of the LmrR structures comprises the β-wing loop (residues 70–75). The drug-binding site is located at the dyad axis of the LmrR dimer. In the LmrR–H33342 complex structure, where the dimer is formed by crystallographic symmetry, the Hoechst compound binds in two mutually exclusive orientations related by the crystallographic dyad symmetry, resulting in an averaged electron density at the drug-binding site (Supplementary Figure S1A). However, electron density for H33342 is well defined, allowing straightforward deconvolution of the two symmetry-related binding modes. Electron density for the daunomycin molecule in the LmrR–daunomycin complex is less well defined (Supplementary Figure S1B), indicating some disorder in binding. A summary of the data collection and model refinement statistics is presented in Table I.
Table 1.
Selected crystallographic data and statistics
| apo–LmrR | LmrR–drug complexes | ||
|---|---|---|---|
| H33342 | Daunomycin | ||
| Data analysis | |||
| Space group | C2221 | P43212 | P212121 |
| Unit cell (Å) | |||
| a | 46.6 | 34.9 | 35.4 |
| b | 52.6 | 34.9 | 53.0 |
| c | 174.9 | 197.0 | 147.1 |
| Resolution (Å) | 35–2.0 | 65–2.2 | 70–2.2 |
| Rmergea | 0.04 (0.5)b | 0.05 (0.206) | 0.036 (0.273) |
| Mean I/σI | 46.6 (2.0) | 21.1 (4.8) | 28.9 (3.8) |
| Completeness (%) | 100 (99.3) | 99.8 (99.0) | 96.0 (99.9) |
| Unique reflections | 15 006 | 6948 | 17 020 |
| Redundancy | 6.7 (7.8) | 3.9 (2.9) | 3.0 (3.5) |
| Refinement statistics | |||
| Resolution (Å) | 23–2.0 | 50–2.2 | 50–2.2 |
| Rwork/Rfreec | 0.22/0.27 | 0.21/0.26 | 0.23/0.27 |
| No. of non-H atoms | |||
| Protein | 1693 | 850 | 1834 |
| Ligand | — | 34 | 38 |
| Waters | 83 | 27 | 43 |
| Root mean square deviations in | |||
| Bond length (Å) | 0.011 | 0.013 | 0.016 |
| Bond angles (deg) | 1.4 | 0.9 | 0.7 |
| Average B-values (Å2) | |||
| Protein | 15 | 21 | 23 |
| Ligand | — | 34 | 73 |
| Ramachandran analysis | |||
| Most favoured (%) | 99.0 | 98.0 | 98.0 |
| Additional allowed (%) | 1.0 | 2.0 | 2.0 |
| aRmerge=ΣΣ∣Ihkl−Ihkl(j)∣/ΣΣIhkl, where Ihkl(j) is the observed intensity and Ihkl is the final average intensity value. | |||
| bValues in parentheses are for the highest resolution shell. | |||
| cRwork=Σ∣∣Fobs∣−∣Fcalc∣∣/Σ∣Fobs∣ and Rfree=Σ∣∣Fobs∣−∣Fcalc∣∣/Σ∣Fobs∣, where all reflections belong to a test set of 10% randomly selected data. | |||
Overall structure of apo–LmrR
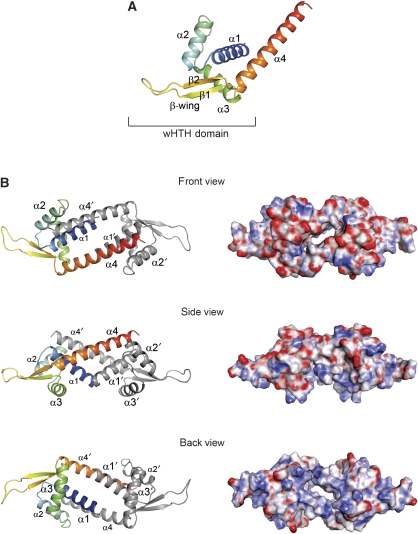
The crystal of apo–LmrR contains two independent copies of a subunit in the asymmetric unit, which each, through a crystallographic dyad rotation axis, forms a biologically relevant dimer with approximate overall dimensions of 100 Å × 38 Å × 38 Å (Figure 1). Each LmrR subunit has an (α+β) structure with topology α1 (residues 6–23), α2 (residues 28–39), α3 (residues 47–60), β1 (residues 63–67), β2 (residues 77–81), α4 (residues 83–108), and is divided into two functional domains: a typical wHTH DNA-binding domain, which consists of helices α1, α2, the DNA recognition helix α3 and strands β1 and β2 (together forming the wing), and a dimerization domain containing the C-terminal helix α4 (Figure 1A). Helix α4 forms a protruding arm, which in the dimer crosses over to the wHTH domain of the dyad-related subunit, packing in a nearly antiparallel orientation against helix α1′ (the prime indicates the other subunit), as well as interacting with the C-terminal region of helix α2′ and the loop connecting helices α2′ and α3′ (Figure 1B). Although facing each other at the centre of the dimer, there is no interaction between the C-terminal helices α4 and α4′, nor between N-terminal helices α1 and α1′. Remarkably, this dimeric arrangement results in the formation of a large flat-shaped pore (approximately 22 Å in width and 6 Å in height) running through the dimer centred around the dyad. The pore entrances are formed by helices α4 and α4′ on one side (hereafter named the front entrance), and by helices α1 and α1′ and the two DNA recognition helices α3 and α3′ on the other side (back entrance). The inside of the pore is largely hydrophobic and formed by residues from the N- and C-terminal helices of both subunits. The pore centre is constricted by a dyad-related pair of tryptophan residues (W96 in α4 and α4′), the indole rings of which are oriented face to face at a distance of about 7 Å (as calculated from the centres of mass of the indole groups). Clusters of arginine and lysine residues surround the back entrance (K6, R10, K55, K59, R75 and K77 from each subunit) resulting in a net positive surface charge, which is consistent with that side of the dimer forming the binding site for DNA. By contrast, the surface around the front entrance is largely negatively charged due to the presence of 12 Glu and Asp residues, 6 from each C-terminal helix in the dimer (E83, E87, E94, D100, E104 and E107). The opposite electrostatic surface charges around the front and back entrances (Figure 1B) create a small but significant overall molecular dipole moment (1192 Debye) running through the pore coinciding with the dyad axis.
Figure 1.
Overall structure of apo–LmrR. (A) Ribbon representation of a single LmrR subunit with a rainbow colour gradient from the N terminus (blue) to the C terminus (red). Secondary structure elements are indicated with labels. (B) The apo–LmrR dimer is shown in three orientations, related by 90° rotations, resulting in a front view (along the two-fold rotation axis facing the α4 helices), a side view (perpendicular to the two-fold axis) and a back view (along the two-fold axis facing the α1 and α3 helices). Helices are indicated with labels. The left panel shows the LmrR dimer in ribbon presentations, the right panel in electrostatic surface representations. The red and blue in the surface representations indicate the strength of the electrostatic surface potential (red, negative charge; blue, positive charge).
The dimer interface
A striking feature of the LmrR structure is its unusual dimeric arrangement leading to the formation of a large central pore. As pointed out above, the dimer interface is formed by interactions of the C-terminal helix of each subunit with the DNA-binding domain of its dimer mate. The surface area of one subunit that becomes buried upon dimerization is ∼1160 Å2, which is within the expected range for a stable dimer considering the 13.5-kDa size of the LmrR subunit (Janin et al, 1988). However, the buried surface area in the LmrR dimer is substantially smaller than the buried surface areas in other MarR/PadR dimers (e.g. the buried surface area in the MarR dimer is 3700 Å2), in which dimerization usually involves more inter-helical interactions and the formation of a central compact core. Stabilization of the LmrR dimer mainly occurs through hydrophobic interactions. Residues in helix α4 that participate in forming the dimer interface are L91, A92, W96, R98, V99, I102, I103, N105 and L106. In the wHTH domain of the dimer-related subunit, the residues important for dimerization are M8′, A11′, Q12′, V15′, I16′ and V20′ (from helix α1′), and V35′, A38′, N40′ and M43′ (from helix α2′ and the loop connecting helices α2′ and α3′). A few interactions are of polar or ionic nature. For example, the side chain of N105 forms a hydrogen bond with the main chain oxygen of A38′, and a salt bridge is formed between R98 and E42′. Another notable residue at the dimer interface is Q12′ in helix α1′. The side chain of this residue forms a hydrogen bond with S95 in helix α4, and makes a π–cation interaction with W96 at the back face of the indole rings (assigning the one exposed towards the pore as their front face), thus stabilizing the conformation of this central residue in the pore.
Comparison with other winged helix proteins
A search of the PDB using the Dali server (Holm and Sander, 1996) showed that the LmrR subunit has significant structural homology with various DNA-binding proteins containing helix-turn-helix or wHTH domains. Among these are the structurally and functionally characterized MarR/PadR family members, MarR of Escherichia coli (PDB accession code 1JGS, 87 equivalent Cα atoms were superimposed with a root mean square deviation (RMSD) of 2.3 Å), OhrR of Bacillus subtilis (1Z9C, 86 equivalent Cα atoms were superimposed with an RMSD of 2.8 Å), AphA of Vibrio cholerae (1YG2, 82 equivalent Cα atoms were superimposed with an RMSD of 3.7 Å) and Pex of Synechococcus sp. (2E1N, 78 equivalent Cα atoms were superimposed with an RMSD of 1.8 Å). The structural similarities of LmrR with the MarR/PadR proteins are mainly confined to its wHTH domain and the sequence identities are rather low (ranging from 11% for MarR to 28% for Pex). Interestingly, structural similarity was also detected with a number of hypothetical transcriptional regulators in the PDB, for which structure and function have not been published. One such protein, from Clostridium thermocellum, showed a particularly high structural homology with LmrR (PDB accession code 1XMA, 99 equivalent Cα atoms were superimposed with an RMSD of 2.7 Å and a sequence identity of 35%). A structure-based sequence alignment of LmrR with 1XMA, AphA, MarR and OhrR is presented in Figure 2A. Most of the conserved residues are hydrophobic and appear to be important in the stabilization of the overall fold of the DNA-binding domain. Currently, the only MarR/PadR protein for which a DNA-bound structure is known is OhrR (Hong et al, 2005). The regions in LmrR that are equivalent to the DNA-binding site in OhrR, that is, helix α3 and the β-wing, show the highest degree of conservation. Nevertheless, among the conserved residues only a few have a role in specific DNA binding in OhrR, suggesting that LmrR and OhrR recognize different DNA sequences. Interestingly, the putative DNA-binding sites of LmrR and 1XMA are highly conserved, which may indicate that these proteins bind similar DNA sequences. Unfortunately, the specific DNA-binding sequence(s) of LmrR within the lmrR and lmrCD promoters have not yet been delineated (Agustiandari et al, 2008), and for the C. thermocellum homologue the target promoters are unknown. The comparison of LmrR with the four MarR/PadR transcription factors further reveals two invariant residues, G61 and G85, which are found in regions that connect the β-wing to helices α3 and α4 and appear to have crucial structural roles in stabilizing the conformation of the β-wing relative to the DNA recognition helix and the dimerization domain. Another invariant residue is T82, which is located at the proximal end of the wing between β2 and helix α4. Previously, it was shown that a T82I mutation in LmrR is associated with drug-resistant phenotypes of L. lactis (Lubelski et al, 2006), and that the LmrR–T82I mutant is deficient in both drug and DNA binding (Agustiandari et al, 2008). In the LmrR structure, the side chain hydroxyl group of T82 makes three hydrogen bonds, with the backbone carbonyl of G61 at the N-terminal end of the β-wing and with the backbone amides of I84 and G85 at the N terminus of helix α4, whereas the main chain amide of T82 forms a hydrogen bond with the backbone carbonyl of I62 (Supplementary Figure S2). These interactions are conserved in the other MarR/PadR structures, and appear to clamp the β-wing in place and stabilize the C-terminal helix. Replacement of T82 by an isoleucine residue will result in the loss of at least three of the four hydrogen bonds, as well as in the introduction of significant steric strain due to the presence of a bulkier and more hydrophobic side chain. Thus, the T82I mutation is predicted to induce a structural perturbation of LmrR, which probably corresponds to a detachment of the β-wing and a disordering of its C-terminal helix. Such a structural perturbation would explain the deleterious effect of the T82I mutation on overall LmrR function.
Figure 2.
Comparison with other wHTH proteins. (A) Structure-based multiple sequence alignment computed with the Tcoffee web server (Poirot et al, 2003) and visualized with JalView (Clamp et al, 2004). Structures were taken from the PDB using the following accession numbers: 1XMA, 1YG2 (AphA), 1JGS (MarR) and 1Z9C (OhrR). Residues of LmrR involved in dimerization are indicated with green dots, residues involved in drug binding with grey dots, and residues in OhrR involved in DNA binding are indicated with blue dots. (B) Ribbon representations of the LmrR and 1XMA dimers. Equivalent secondary structure elements are indicated in specific colors: α1, blue; α2, grey; α3, magenta; α4, red and the β-wing, green.
The structural organization of the LmrR dimer, with the two DNA-binding domains adjacent to one another and their DNA recognition helices facing the same side, is similar to other MarR/PadR family members, pointing to a similar mode of DNA binding with the recognition helices fitting into two successive major grooves on one side of the DNA double helix. However, the dimeric architecture of LmrR is unique: none of the other structurally characterized MarR/PadR family members shows a central pore at the dimer interface. It should be noted, though, that the dimerization modules in MarR/PadR proteins are highly diverse, consisting of different numbers of α-helices packed together in various ways. Of the four proteins that were compared with LmrR, only 1XMA has an identical overall topology containing a single C-terminal dimerization helix. Interestingly, the sequence homology of LmrR with 1XMA extends into the C-terminal helix and among the conserved residues are W96, as well as residues near the C terminus that in LmrR participate in dimer formation. Nevertheless, unlike in LmrR, in the C. thermocellum transcription factor the N- and C-terminal helices of the two subunits form a compact core at the dimeric interface that is completely closed. This difference at the dimer interface is coupled to a difference in the relative orientation of the C-terminal helices with respect to the wHTH domains and to significant bending of the N- and C-terminal helices (Figure 2B), which allow the subunits in the dimer of the C. thermocellum transcription factor to approach each other more closely than the subunits in the LmrR dimer. To predict whether the ‘closed' conformation of the 1XMA dimer would be accessible to LmrR, a 1XMA-based homology model was prepared of LmrR using the SWISS model server (http://swissmodel.expasy.org/). The results indicate that there are no major steric clashes that would prevent LmrR to adopt a ‘closed' conformation. However, in comparison with the 1XMA dimer, the conserved tryptophan pair and their surrounding residues in LmrR seem unsuitable to provide the necessary hydrophobic packing and interactions to stabilize a closure of the central pore (Supplementary Figure S3). On the other hand, additional overall conformation changes and rearrangements of side chains could perhaps create a well-packed hydrophobic core. Clearly, homology modelling alone is not sufficient to assess the likelihood of LmrR also adopting a ‘closed' conformation. More sophisticated methods, such as molecular dynamic simulations or NMR, could perhaps provide an answer, but these are beyond the scope of the present study.
Binding of H33342 and daunomycin
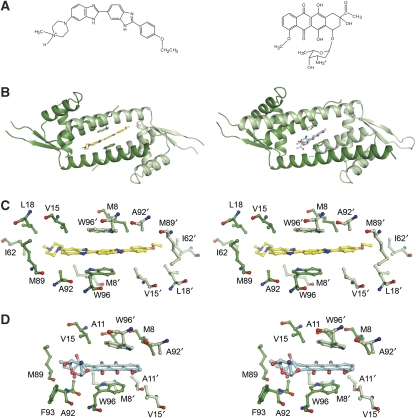
The structures of LmrR bound with H33342 and daunomycin reveal that the central pore in the LmrR dimer serves as a multidrug-binding site (Figure 3). In both complexes, the pore accommodates a single drug molecule, consistent with the 1:2 (drug:LmrR subunit) stoichiometry of drug binding obtained previously from fluorescent titrations with H33342 (Agustiandari et al, 2008). The two drugs show a common mode of binding: their flat ring systems are wedged in between the W96 and W96′ side chains forming aromatic stacking interactions with each of the two indole systems (Figure 3C and D). Further stabilization of the bound drugs is provided by various hydrophobic contacts in the pore. Remarkably, no hydrogen bonds are observed between the protein and the drugs. Rather, the orientation of the drugs is such that most of their polar atoms are in a solvent-exposed position facing the front or back entrances of the pore. Several bound water molecules are present in the pore to function as hydrogen bond partners of the drugs.
Figure 3.
Crystallographic analysis of drug binding to LmrR. (A) Chemical structures of H33342 (left) and daunomycin (right). (B) Ribbon diagrams of the LmrR–H33342 (left) and LmrR–daunomycin (right) complex, showing the drug molecule (sticks) bound inside the central pore of the dimer in between the two tryptophan residues W96 and W96′ (sticks). The two subunits of the LmrR dimer, as well as W96 and W96′, are coloured with different shades of green. (C) Close-up stereo view of the drug-binding pore in the LmrR–H33342 complex depicting the drug and residues that contact the drug (maximum contact distance defined as 4 Å). (D) Similar stereo view as in (C) of the drug-binding pore in the LmrR–daunomycin complex.
The two drug-bound LmrR structures also show substantial differences. The elongated crescent-shaped Hoechst compound deeply penetrates the pore and stretches out over its entire width with the ethoxy phenolic and N-methyl piperazine groups extending towards the sidewalls and facing the back entrance of the pore. The area of the drug–protein interaction surface is substantial (∼215 Å2) and shows good shape complementarity. W96 and W96′ clamp down one of the central benzimidazole ring systems, which are aligned in an off-centered parallel orientation with respect to each of the two indole rings. The inter-ring distances are ∼3.5 Å, optimal for allowing the formation of strong van der Waals interactions (McGaughey et al, 1998).
Less extensive interactions are formed with daunomycin. The presence of the bulky amino sugar substituent prohibits a deep penetration of the drug into the flat pore (Figure 5C). Only the aglycon chromophore interacts with the protein, whereas the amino sugar is exposed to the solvent at the front entrance of the pore. As a consequence, the area of the drug–protein interaction surface in the LmrR–daunomycin complex is much smaller (∼82 Å2) than in the LmrR–H33342 complex. No electron density could be observed for the amino sugar of daunomycin, indicating that this substituent is highly flexible. The stacking interactions of the aromatic rings with the W96/W96′ pair form the main contribution to drug binding stabilization, but the stacking geometry is less optimal than in the LmrR–H33342 complex. The weaker interactions between LmrR and daunomycin, as compared with the interactions between LmrR and H33342, are further illustrated by the relatively high atomic B-factors of daunomycin (Table I).
Figure 5.
Conformational differences between the apo and two drug-bound structures of LmrR. (A) Superposition (in ribbon representation) of the apo–LmrR subunit structure (yellow) and the subunit structures of H33342-bound (green) and daunomycin-bound LmrR (salmon). The superposition was carried out using the Cα atoms of the wHTH domain. (B) Superposition of the three LmrR dimers, showing the difference in relative position of the two wHTH domains. Only one of the two subunits (light colors) was used for the superposition (identical to the superposition in Figure 5A). The range of distances between the two DNA recognition helices in the different dimers, and the largest rotational shift of the wHTH domains (based on comparing the H33342- and daunomycin-bound dimers) are indicated. (C) The same superposition as in Figure 5B, but from a different view, showing the relative shifts of helix pair α1–α4′ with respect to helix pair α1′–α4. H33342 (green) and daunomycin (salmon), as well as the W96/W96′ tryptophan pair, are also shown in sticks. The amino sugar moiety of daunomycin is shown in a solvent-exposed position at the front entrance of the pore, but it should be noted that its binding is highly disordered, as evident from the weak electron density associated with this substituent.
Drug-binding affinities and importance of W96 for drug and DNA binding
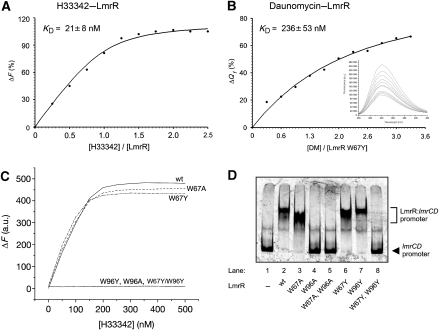
To quantify the difference in binding affinity of LmrR for H33342 and daunomycin, the dissociation constants of the two drugs were approximated from binding curves obtained from two different fluorescence-based drug-binding assays (Figure 4A and B). Binding of H33342 to untagged LmrR was monitored by recording the increase in drug fluorescence when H33342 moves from an aqueous to a hydrophobic environment, that is, when it binds to the drug-binding site of LmrR. The apparent dissociation constant of H33342 obtained from fitting the binding curve was ∼20 nM, showing that this compound has a strong affinity for the drug-binding site of LmrR. Unfortunately, the spectral properties of daunomycin did not allow the use of a similar binding assay. Instead for daunomycin, we obtained a binding curve by measuring the fluorescence quenching of W96 upon titration of the drug. LmrR contains two tryptophan residues, W67 and W96, the former of which is located in the β-wing of the DNA-binding domain. To avoid unwanted disturbances of the fluorescence signal, W67 was mutated to either alanine or tyrosine. The W67Y and W67A mutations had no significant effect on the binding of H33342 by LmrR, nor on the binding of lmrCD promoter DNA (Figure 4C and D). The dissociation constant of daunomycin, determined from W96 fluorescence quenching using the LmrR–W67Y mutant, was ∼0.25 μM, confirming that its binding affinity for LmrR is weaker than the Hoechst compound.
Figure 4.
Spectroscopic and mutational analysis of drug and DNA binding by LmrR. (A, B) Fluorescence titration curves measuring H33342 and daunomycin binding to LmrR. Binding of H33342 was monitored by recording the increase in drug fluorescence when titrating a solution of wild-type LmrR with increasing concentrations of H33342, using the experimental conditions described (Agustiandari et al, 2008). Binding of daunomycin was monitored by performing tryptophan fluorescence quenching titration of the LmrR mutant W67Y with increasing concentrations of daunomycin. The apparent KD values are calculated from the fitting of the data using nonlinear regression analysis (r2=0.993 and 0.996 for H33342 and daunomycin, respectively). Inset, tryptophan fluorescence emission spectra in the presence of increasing concentrations of daunomycin. (C) Fluorescence titration curves measuring H33342 binding to the W67 and W97 single mutants and the W67Y/W96Y double mutant of LmrR. The binding curves for the W96 single mutants, as well as the W67A/W96Y double mutant, are flat, thus revealing that these mutants lack drug-binding capability. (D) Electrophoretic mobility shift assay (EMSA) of the LmrR mutants analysing their binding to the lmrCD promotor DNA.
To confirm its importance for drug binding, W96 was also mutated to alanine or tyrosine. Both these LmrR mutants, W96A and W96Y, lost the ability to bind H33342 (Figure 4C). Interestingly, although the W96Y mutant was still able to bind to the lmrCD promotor, the W96A mutant was not, nor was a W67Y/W96Y double mutant (Figure 4D). The impaired DNA binding of the W67Y/W96Y mutant is not due to a loss of structural integrity of the protein, as was assessed by circular dichroism (Supplementary Figure S4A). These results thus point to some indirect role in DNA binding for residue 96, in addition to a direct role in drug binding.
Conformational flexibility
As pointed out earlier, notable conformational differences were identified between the different LmrR structures (Figure 5). Pairwise structural superpositions of the isolated subunits in the different dimers, using all Cα atoms, result in RMSDs ranging from 1.0 to 2.0 Å. These deviations primarily result from differences in the orientation of the C-terminal helix α4 relative to the wHTH domain. The reorientations of helix α4 may be described as lever-arm rotations with the residues that attach the N terminus of the helix to the wHTH domain serving as a hinge (Figure 5A). Even though the rotations are small (varying between 9° and 16°), the lever-like movement results in substantial translational shifts of residues near the C terminus of helix α4 (up to ∼8 Å as calculated from Cα–Cα distances). Except for small shifts of helix α1 and α2, no significant conformational changes are observed in the wHTH domains. Through the dimeric interface, the conformational differences in the subunits are coupled to two pronounced differences in the LmrR dimeric structures. First, when comparing the H33342- and daunomycin-bound LmrR dimers with the apo–LmrR dimer, one of the wHTH domains is rotated relative to the wHTH domain of the other subunit. The amounts of rotation are about equal for both drug-bound complexes (∼14°), but the rotations are in opposite directions (Figure 5B). In fact, the largest conformational change (∼28° rotation) is observed when mutually comparing the H33342- and daunomycin-bound LmrR dimers. The rotations of the wHTH domains are coupled to changes in the spacing between the two DNA recognition helices. The shortest spacing is observed in the daunomycin-bound complex with a distance of ∼26 Å (measured from the centroids of the helices), whereas in the H33342-bound complex the spacing is the largest with a distance of ∼32 Å. Second, inside the pore, differences are observed in the orientations of residues from one subunit relative to those of the other subunit. These differences are caused by shifts of the α1–α4′ helix pair relatively to the α1′–α4 helix pair in the different LmrR structures, and directly affect the geometry of the central drug-binding site (Figure 5C).
Unfortunately, as each crystal structure of LmrR represents a different crystal form, it is not possible to distinguish whether the conformational changes are drug induced, or whether they are caused by differences in crystal packing. However, the observed structural differences point to a remarkable plasticity of LmrR. As the conformational rearrangements affect both the drug binding and DNA-binding sites in the LmrR dimer, similar conformational changes most likely have an important function in the induction mechanism of LmrR.
Discussion
The crystal structure of LmrR is the first structure of a transcription factor regulating the expression of a multidrug ABC transporter. It is also the first time that the structural basis of multidrug recognition has been studied for a PadR transcriptional regulator. Current knowledge of the mechanism of multidrug recognition by transcriptional regulators of multidrug transporters is largely based on crystallographic studies with the transcription factors BmrR from B. subtilis (Zheleznova et al, 1999), QacR from Staphylococcus aureus (Schumacher et al, 2001; Schumacher and Brennan, 2003) and TtgR from Pseudomonas putida (Alguel et al, 2007). The structures of LmrR bound to H33342 and daunomycin confirm the importance of several of the general structural features that seem important in multidrug recognition, for example, the availability of a large drug-binding pocket that can accommodate a large spectrum of drug ligands, the importance of aromatic and hydrophobic residues for providing van der Waals interactions to stabilize the bound ligands, the importance of water molecules for occupying regions of the pocket not occupied by the ligand and for solvating hydrophilic groups of the ligand that do not interact with the protein, and a flexible pocket wall that can change conformation upon ligand binding. However, there are also some striking differences in the way multidrug binding is accomplished by LmrR as compared with BmrR and QacR. The foremost difference is that in LmrR the multidrug-binding pocket is formed by a symmetric pore located at the dimer centre with both subunits contributing equally to its architecture. In QacR and BmrR, the drug-binding pockets are asymmetric and primarily formed within a single subunit. Also, in the drug-bound LmrR complexes the binding modes of the two different drugs are very similar and involve a common and strong aromatic stacking interaction with the W96/W96′ tryptophan pair. In the other transcription factors, the architecture of the drug-binding pockets allows different ligands to adopt different orientations within the pocket and to interact with different sets of amino acids. In BmrR and QacR, the binding affinity for cationic drugs is further augmented by electrostatic attraction between the positively charged ligand and buried or partially buried negatively charged glutamates or aspartate residues of the protein. No such interactions were observed in the H33342- and daunomycin-bound LmrR structures. However, in LmrR, positive charges in the drugs may be stabilized by long-range electrostatic interactions with the cluster of glutamate and aspartate residues that surround the front entrance of the pore. Also, the apparent molecular dipole moment that was found running through the pore of LmrR may assist in attracting and binding cationic drugs. The relevance of this latter feature of the LmrR structure for drug specificity is, however, very unclear: it could be likewise important for directing LmrR towards the DNA substrate.
It is evident that the homology of LmrR with other members of the PadR protein family that have so far been functionally and structurally characterized, that is, AphA and Pex, is relatively low. Our results thus confirm the classification of two PadR subfamilies (Huillet et al, 2006): subfamily I with longer sequences (∼180 amino acids) to which AphA and Pex belong, and a more distant subfamily II with shorter sequences (∼110 amino acids) to which LmrR belongs. To the best of our knowledge, LmrR is so far the only member of PadR subfamily II that has been characterized both functionally and structurally. Interestingly, a BLAST search against translated nucleotide sequence databases (see Supplementary Figure S5) yields a large number of close homologues of LmrR that exist in various bacterial species, in particular those belonging to the Firmicutes (Listeria, Bacillus, Staphylococcus, Enterococcus, Lactobacillus, Streptococcus and Clostridium). Both the N- and C-terminal domains of LmrR are significantly conserved in these proteins, including W96, thus pointing to a high similarity in overall structure and dimeric organization. Most likely, some of these proteins also have a similar role as LmrR in regulating multidrug resistance, but lack of functional data prohibits such assessment for the moment. Furthermore, the sequence conservation in the C-terminal helix of these LmrR-like proteins is no guarantee for the existence of a central multidrug-binding pore, as is evident from the structural comparison of LmrR with the C. thermocellum homologue. Future studies should therefore reveal whether the multidrug binding characteristics of LmrR are applicable to a larger set of proteins.
In the absence of a DNA-bound structure, the induction mechanism of LmrR remains to be determined. Comparison of the different LmrR structures, and the effects of the W96 mutations, reveals a possible allosteric coupling between the drug- and DNA-binding sites. Most likely the binding of a drug to LmrR locks the dimer in a conformational state that is incompatible with DNA binding, due to a relative positioning of the DNA recognition helices that is unsuitable for their simultaneous insertion in the successive major grooves of the DNA. Such an induction mechanism would be similar to the induction mechanism of various wHTH domains containing transcription factors, although the origin and nature of the structural changes involved in this mechanism are most likely to be different for LmrR. To explore this further, we tested the suitability of the different LmrR structures to bind B-form DNA using model building. None of the three LmrR structures has a conformation that allows a good fit with DNA (Supplementary Figure S6). In daunomycin-bound LmrR, the DNA recognition helices are spaced too close together and would sterically clash with the DNA. In contrast, in the apo form and H33342-bound complex of LmrR, they are positioned too far apart, and one of the two wHTH DNA-binding domains is shifted away from the DNA. It should be noted, though, that such modelling does not take into account the possibility of DNA distortion, thus limiting its significance. Further studies are underway to better define the DNA-binding characteristics of LmrR and unequivocally identify its operator DNA sequence. This knowledge will be crucial to allow crystallization of a DNA-bound LmrR complex.
Materials and methods
Chemicals
H33342 (Molecular Probes) and daunomycin (Calbiochem) were purchased and used without further purifications.
Protein production, crystallization and X-ray data collection
Tagged LmrR, comprising full-length LmrR (from L. lactis strain MG1363) and a C-terminal streptactin tag (117-SRWSHPQFEK-126), was obtained by nisin-induced overexpression in L. lactis using the expression vector pNSC8048-lmrR, and initially purified through streptactin-tag affinity and heparin column chromatography as described elsewhere (Agustiandari et al, 2008). Final purification to homogeneity was achieved by size-exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare) with a running buffer containing 20 mM Tris–HCl, pH 8.0, 280 mM NaCl and 1 mM EDTA. To anticipate possible negative effects of the streptactin tag on protein crystal growth or protein conformation, untagged full-length LmrR was also produced using a modified version of the overexpression plasmid in which the streptactin tag encoding sequence was deleted. Untagged LmrR was first purified on a heparin column with conditions similar to those used for tagged LmrR, followed by purification on a Mono S HR 5/5 cation exchange column (GE Healthcare) with a linear gradient of 0.03–1 M NaCl (20 mM Hepes, pH 8, 1 mM EDTA, 0.5 mM DTT). Finally, the protein was loaded on a Superdex 200 10/300 GL column and eluted using the same running buffer as for tagged LmrR.
Purified LmrR, with or without streptactin tag, was concentrated in the gel filtration running buffer and either used immediately for crystallization or frozen in liquid nitrogen and stored at −80°C. Crystallization trials were set up with the aid of an Oryx6 crystallization robot (Douglas Instruments) using the PACT and JCSG crystallization screens (Newman et al, 2005). Lead conditions were optimized manually using the sitting drop vapour diffusion method with crystallization drops containing 1 μl of the protein solution (8 mg/ml) and 1 μl of the reservoir solution. Crystals of drug-bound LmrR complexes were obtained by co-crystallization using an LmrR solution (8 mg/ml) preincubated for 30 min with 2 mM H33342 or daunomycin. Diffracting crystals of LmrR in drug-free conditions were obtained with both tagged and untagged proteins, whereas in the drug-bound form only streptactin-tagged LmrR yielded well diffracting crystals. At drug-free conditions, tagged LmrR crystals were obtained with 20% PEG 3350 in 0.1 M Bis–Tris propane, pH 8.5 and 0.2 M NaNO3, whereas drug-free untagged LmrR crystals grew in 30% PEG 1500, 0.1 M propionic acid/cacodylate/Bis–Tris propane cocktail buffer, pH 8.5. Crystals of H33342-bound LmrR grew against a well solution containing 25% PEG 1500, 0.1 M succinic acid/phosphate/glycine buffer, pH 9.0, whereas crystals of daunomycin-bound LmrR–strep were obtained with 25% PEG 1500, 0.1 M malonic acid/imidazol/boric buffer, pH 7.0. All crystals grew overnight at room temperature.
X-ray diffraction data were collected at cryogenic temperatures by using the MX beam lines of the European Synchrotron Research Facility (ESRF) at Grenoble. Prior to data collection, crystals were flash cooled in a cryoprotectant solution of mother liquor with 20% glycerol. The data were processed with MOSFLM (Leslie, 2006) and merged using SCALA as implemented in CCP4. Relevant data statistics are shown in Table I. Data on the tagged LmrR crystal grown in the absence of drugs, and its derived structure, will not be presented here, as they were merely used to verify that the C-terminal streptactin tag did not affect the overall LmrR structure.
Structure determination of apo–LmrR
In the absence of drugs, untagged LmrR was crystallized in the space group C2221 with two subunits of two different dimers in the asymmetric unit. The crystals diffracted up to 2.0-Å resolution. The untagged apo–LmrR structure was solved by molecular replacement using PHASER with the automated search process (McCoy, 2007). Various search models were prepared and tried using the structures of homologous proteins from the PDB, as identified by the FFAS server (Jaroszewski et al, 2005). Molecular replacement succeeded with a search ensemble containing the structures of three hypothetical transcription factors (PDB entries 2ESH, 1YYV and 1XMA) having sequence identities with LmrR ranging from 15 to 34%. Phase improvement and construction of an initial protein model was performed by using the automatic map improvement and model building routines in RESOLVE (Terwilliger, 2003). The final model was obtained by carrying out various cycles of refinement using REFMAC (Murshudov et al, 1997) interspersed with cycles of rebuilding and placement of water molecules using COOT (Emsley and Cowtan, 2004). TLS refinement was used in the last refinement cycles to model anisotropic displacements (Winn et al, 2001, 2003). The final model of the apo–LmrR structure contains two polypeptide chains: one discontinuous chain covering residues 5–70 and 75–109, and one continuous chain covering residues 5–109. In both polypeptides, residues 1–4 and 110–116 are missing due to weak or absent electron density. Each polypeptide is one subunit of different biological dimers that are formed by crystallographic two-fold axes.
Structure determination of drug-bound LmrR
H33342-bound LmrR crystals belong to the space group P43212 with one molecule per asymmetric unit, whereas daunomycin-bound LmrR crystallized in a different space group (P212121) with a dimer in the asymmetric unit. The structures of the drug-bound complexes were solved by molecular replacement using the apo–LmrR monomer as a search model. Clear density in 2Fo−Fc and Fo−Fc Fourier maps, calculated at the initial stages of refinement, indicated the location and binding mode of the drugs. The model building and refinement were done with COOT and REFMAC5. In both cases, TLS refinement was used in the last refinement cycles. For the LmrR–H33342 complex, the final protein model contains residues 3–108. No electron density is observed for residues 1–2, 109–126 (including the streptactin tag) and 71–73 (β-wing) loop. The final protein model of LmrR–daunomycin contains residues 1–70 and 73–116 for chain A, and residues 5–115 for chain B.
Structure analysis
Relevant crystallographic statistics of the refined models are shown in Table I. Stereochemistry of the models was validated with the programs Procheck (Laskowski et al, 1993) and MolProbity (Davis et al, 2007). 3D structural superpositions and assessment of conformational differences were carried out with the programs Lsqman (Kleywegt, 1999) and Dyndom (Hayward and Berendsen, 1998). Electrostatic surface potentials were calculated using APBS (Baker et al, 2001) and visualized using PyMOL (Delano Scientific). The molecular dipole moment of the LmrR dimer was calculated using the protein dipole moments server at http://bioportal.weizmann.ac.il/dipol. Additional analyses, such as calculation of surface areas, were performed with various programs from the CCP4 program suite.
Site-directed mutagenesis
Mutations of W67 and W96 in the lmrR gene were performed through round PCR using the pNSC8048-lmrR plasmid as the template together with synthetic primers containing the designated mutations. PCR products were ligated at 4°C overnight before being transformed to the L. lactis NZ9000-competent cells through electroporation. Selected colonies were inoculated at 30°C in M17 media (Difco) supplemented with 0.5% glucose (w/v) and 5 μg/ml chloroamphenicol. Plasmid isolation was performed using GenElute Plasmid miniprep kit (Sigma-Aldrich) and the correct mutations were verified through nucleotide sequencing.
Drug-binding assays
Binding of H33342 to the DNA-free purified LmrR variants was monitored by the increase of H33342 fluorescence upon binding as described earlier (Agustiandari et al, 2008). Binding of daunomycin to LmrR mutant W67Y was monitored by tryptophan fluorescence quenching titration experiments using an Aminco Bowman Series 2 spectrofluorometer (excitation wavelength of 295 nm, emission spectra obtained from 300 to 450 nm). A detailed description of the drug-binding assays, additional control experiments and the procedure that was followed to derive the apparent dissociation constants (KD) of the two drugs is included in the Supplementary data.
DNA-binding assay
The ability of the LmrR variants to bind to a 287-bp fragment corresponding to the promoter region of lmrCD was studied by means of an electrophoretic mobility shift assay as described (Agustiandari et al, 2008).
Circular dichroism
Circular dichroism spectra were obtained at 25°C by using an Aviv 62ADS spectropolarimeter (Aviv Associates, Lakewood, NJ). The protein samples contained 0.24 mg/ml protein in 20 mM Tris–HCl, pH 8.0 and 50 mM NaCl.
Accession numbers
The atomic coordinates and structure factors for apo–LmrR (entry 3F8B), H33342-bound LmrR (entry 3F8C) and daunomycin-bound LmrR (entry 3F8F) have been deposited in the PDB, Research Collaboratory for Structural Bioinformatics, Rutgers University (http://www.rcsb.org).
Supplementary Material
Supplementary Figures S1 and S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary data
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and we thank the MX beamline scientists for assistance in beamline usage.
References
- Agustiandari H, Lubelski J, van den Berg van Saparoea HB, Kuipers OP, Driessen AJ (2008) LmrR is a transcriptional repressor of expression of the multidrug ABC transporter LmrCD in Lactococcus lactis. J Bacteriol 190: 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF (2001) The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat Struct Biol 8: 710–714 [DOI] [PubMed] [Google Scholar]
- Alguel Y, Meng C, Terán W, Krell T, Ramos JL, Gallegos M-T, Zhang X (2007) Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials. J Mol Biol 369: 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29: 231–262 [DOI] [PubMed] [Google Scholar]
- Arita K, Hashimoto H, Igari K, Akaboshi M, Kutsuna S, Sato M, Shimizu T (2007) Structural and biochemical characterization of a cyanobacterium circadian clock-modifier protein. J Biol Chem 282: 1128–1135 [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20: 426–427 [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ (2005) Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J Biol Chem 280: 13779–13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gury J, Barthelmebs L, Tran NP, Divies C, Cavin JF (2004) Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl Environ Microbiol 70: 2146–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S, Berendsen HJ (1998) Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30: 144–154 [PubMed] [Google Scholar]
- Heldwein EE, Brennan RG (2001) Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409: 378–382 [DOI] [PubMed] [Google Scholar]
- Higgins CF (2007) Multiple molecular mechanisms for multidrug resistance transporters. Nature 446: 749–757 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1996) Mapping the protein universe. Science 273: 595–603 [DOI] [PubMed] [Google Scholar]
- Hong M, Fuangthong M, Helmann JD, Brennan RG (2005) Structure of an OhrR–ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell 20: 131–141 [DOI] [PubMed] [Google Scholar]
- Huillet E, Velge P, Vallaeys T, Pardon P (2006) LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol Lett 254: 87–94 [DOI] [PubMed] [Google Scholar]
- Janin J, Miller S, Chothia C (1988) Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol 204: 155–164 [DOI] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A (2005) FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Res 33: W284–W288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ (1999) Experimental assessment of differences between related protein crystal structures. Acta Crystallogr D Biol Crystallogr 55: 1878–1884 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26: 283–291 [Google Scholar]
- Leslie AG (2006) The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr 62: 48–57 [DOI] [PubMed] [Google Scholar]
- Lubelski J, de Jong A, van Merkerk R, Agustiandari H, Kuipers OP, Kok J, Driessen AJ (2006) LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol Microbiol 61: 771–781 [DOI] [PubMed] [Google Scholar]
- McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey GB, Gagne M, Rappe AK (1998) pi-Stacking interactions. Alive and well in proteins. J Biol Chem 273: 15458–15463 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Newman J, Egan D, Walter TS, Meged R, Berry I, Ben Jelloul M, Sussman JL, Stuart DI, Perrakis A (2005) Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: the PACT/JCSG+ strategy. Acta Crystallogr D Biol Crystallogr 61: 1426–1431 [DOI] [PubMed] [Google Scholar]
- Poirot O, O'Toole E, Notredame C (2003) Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res 31: 3503–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr, Paulsen IT, Sliwinski MK, Pao SS, Skurray RA, Nikaido H (1998) Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J 12: 265–274 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Brennan RG (2003) Deciphering the molecular basis of multidrug recognition: crystal structures of the Staphylococcus aureus multidrug binding transcription regulator QacR. Res Microbiol 154: 69–77 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG (2001) Structural mechanisms of QacR induction and multidrug recognition. Science 294: 2158–2163 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol 374: 22–37 [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol 374: 300–321 [DOI] [PubMed] [Google Scholar]
- Zheleznova EE, Markham PN, Neyfakh AA, Brennan RG (1999) Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell 96: 353–362 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1 and S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary data