Abstract
Sre1, the fission yeast sterol regulatory element-binding protein, is an ER membrane-bound transcription factor that controls adaptation to low oxygen growth. Under low oxygen, Sre1 is proteolytically cleaved and the N-terminal transcription factor domain (Sre1N) is released from the membrane and enters the nucleus to activate hypoxic gene expression. Ofd1, a prolyl 4-hydroxylase-like 2-oxoglutarate dioxygenase, controls the oxygen-dependent stability of Sre1N. In the presence of oxygen, Ofd1 accelerates the degradation of Sre1N, but under low oxygen Ofd1 is inhibited and Sre1N accumulates. To identify the regulators of Sre1N, we performed a plasmid-based screen for genes that increased Sre1N transcriptional activity. Here, we identify Nro1 (SPCC4B3.07) as a positive regulator of Sre1N stability and a direct inhibitor of Ofd1. In the absence of oxygen, Nro1 binds to the Ofd1 C-terminal degradation domain and inhibits Sre1N degradation. In the presence of oxygen, Nro1 binding to Ofd1 is disrupted, leading to rapid degradation of Sre1N. We conclude that the Ofd1 dioxygenase domain functions as an oxygen sensor that regulates binding of Nro1 to Ofd1 to control oxygen-dependent Sre1N stability.
Keywords: degradation, hypoxia, prolyl hydroxylase, SREBP, yeast
Introduction
To survive, organisms require mechanisms that allow them to adapt to changes in their environment. Cells have developed mechanisms to sense and adapt to fluctuating oxygen concentrations and hypoxic stress. In both mammals and yeast, transcription factors activate a programme of gene expression under low oxygen to promote adaptive metabolic changes that allow growth and survival (Kwast et al, 1998; Emerling and Chandel, 2005). In mammalian cells, 2-oxoglutarate (OG)–Fe(II) dioxygenase family proteins function as oxygen sensors to regulate the hypoxia-inducible factor (HIF) (Ozer and Bruick, 2007). The HIF transcription factor is a master regulator of hypoxic gene expression and is a heterodimer composed of one of three α-subunits (HIF-1α, HIF-2α and HIF-3α) and HIF-1β (Schofield and Ratcliffe, 2005; Gordan and Simon, 2007). Degradation of the HIF-α subunits is controlled by oxygen-dependent, post-translational hydroxyl modifications. In the presence of oxygen, a family of three 2-OG–Fe(II) dioxygenases, PHD1–3, hydroxylate proline residues on HIF-α and target this modified subunit for proteasomal degradation (Dann and Bruick, 2005; Kaelin, 2005). Conversely, hypoxic conditions inhibit the activity of the PHDs, leading to the accumulation of HIF-α and expression of genes required for hypoxic growth.
In the fission yeast Schizosaccharomyces pombe, Sre1 is an endoplasmic reticulum, membrane-bound transcription factor that is a principal activator of hypoxic gene expression (Hughes et al, 2005; Todd et al, 2006). Sre1 is the homologue of the mammalian sterol regulatory element-binding protein (SREBP) that controls cellular cholesterol homoeostasis (Goldstein et al, 2006; Espenshade and Hughes, 2007). Under low oxygen, Sre1 is proteolytically cleaved and the N-terminal transcription factor (Sre1N) enters the nucleus and upregulates genes essential for low oxygen growth (Figure 1A; Hughes et al, 2005, 2007).
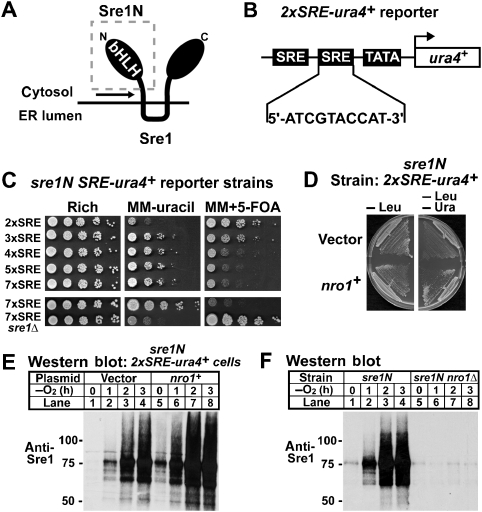
Figure 1.
nro1+ is a positive regulator of Sre1N. (A) Sre1N is the soluble N terminus of Sre1. (B) Diagram of the integrated 2xSRE-ura4+ reporter gene in the sre1N reporter strain. Two tandem SRE sequences fused to a minimal promoter drive the expression of ura4+. (C) sre1N SRE-ura4+ reporter strains with the indicated number of tandem SREs driving ura4+ and sre1Δ 7xSRE-ura4+ cells were spotted in five-fold serial dilutions on rich medium, minimal medium lacking uracil or minimal medium containing 5-FOA. (D) sre1N 2xSRE-ura4+ cells containing empty vector or a plasmid expressing nro1+ from the adh promoter were plated on a minimal medium lacking leucine or lacking leucine and uracil. (E) sre1N 2xSRE-ura4+ cells containing empty vector or a plasmid expressing nro1+ from the adh promoter were grown in the absence of oxygen for the indicated times. (F) sre1N and sre1N nro1Δ cells were grown in the absence of oxygen for the indicated times. For western blotting, whole-cell extracts (40 μg) were subjected to western blot analysis using anti-Sre1 antibody. Molecular mass markers (kDa) are given to left.
Recently, we demonstrated that oxygen acts at a second regulatory point to control the levels of Sre1N (Hughes and Espenshade, 2008). In addition to regulating proteolytic cleavage of Sre1, oxygen controls the stability of Sre1N, such that Sre1N accumulates under low oxygen, but it is rapidly degraded in the presence of oxygen. The prolyl 4-hydroxylase-like, 2-OG–Fe(II) dioxygenase Ofd1 accelerates Sre1N degradation in the presence of oxygen (Hughes and Espenshade, 2008). Ofd1 consists of two domains: an N-terminal 2-OG–Fe(II) dioxygenase domain and a C-terminal degradation domain (CTDD). Interestingly, unlike the HIF prolyl hydroxylases, dioxygenase activity is not required for Sre1N turnover as the Ofd1 CTDD is sufficient to accelerate Sre1N degradation. Rather, the N-terminal dioxygenase domain functions as an oxygen sensor and regulates the ability of the Ofd1 CTDD to destabilize Sre1N. In the absence of oxygen, the N-terminal dioxygenase domain inhibits the Ofd1 CTDD, leading to the accumulation of Sre1N. Conversely, in the presence of oxygen, inhibition is released and Sre1N is rapidly degraded (Hughes and Espenshade, 2008). How activity of the Ofd1 CTDD is regulated by oxygen is unknown.
In this study, we used a growth selection to screen a plasmid cDNA library for positive regulators of Sre1N. We identified Nro1 as a positive regulator of Sre1N stability. Genetic and biochemical experiments demonstrate that Nro1 functions as a direct inhibitor of Ofd1. Nro1 is required for the low oxygen upregulation of Sre1N, and Nro1 suppresses the ability of the Ofd1 CTDD to accelerate Sre1N degradation. In the absence of oxygen, Nro1 binds Ofd1 and inhibits the Ofd1 CTDD, leading to Sre1N accumulation. In the presence of oxygen, Nro1 binding to Ofd1 is disrupted and Sre1N is rapidly degraded. Our studies indicate that the Ofd1 dioxygenase domain functions as an oxygen sensor to control the inhibitory binding of Nro1 to the Ofd1 CTDD and regulate Sre1N stability.
Results
High-copy suppressor screen for positive regulators of Sre1N
Sre1 is activated by proteolytic cleavage to release the soluble N-terminal transcription factor domain Sre1N (Figure 1A). Previous studies demonstrated that degradation of Sre1N was rapid (t1/2=7 min) and that oxygen-dependent degradation required Ofd1 (Hughes and Espenshade, 2008). To identify the regulators of Sre1N activity, we developed a series of integrated reporter constructs that conferred Sre1-dependent growth to fission yeast. The reporter gene consisted of different numbers of the Sre1 DNA-binding site (SRE) from the fission yeast transposon Tf2-1 long terminal repeat, placed upstream of a minimal promoter (Sehgal et al, 2007). This Sre1-dependent promoter directed the expression of ura4+, which codes for orotidine 5-monophosphate decarboxylase (OMPDC) required for uracil biosynthesis and growth on a medium lacking uracil (Figure 1B; Hughes et al, 2008). In addition, OMPDC can convert 5-fluoroorotic acid (5-FOA) to the toxic compound 5-fluorouracil, and cells with high levels of ura4+ expression fail to grow on a medium containing 5-FOA (Boeke et al, 1987).
We generated strains designated sre1N SRE-ura4+ that express Sre1N (Sre1 aa 1–440) from the endogenous sre1+ locus and carry the reporter gene with tandem copies of the SRE sequence (Hughes et al, 2008). Sre1N expression results from the insertion of a termination codon prior to the first transmembrane segment of Sre1 (Figure 1A). Thus, expression of Sre1N does not require proteolytic cleavage, and Sre1N is expressed in the presence of oxygen. To assay the ability of the SREs to direct the expression of the ura4+ reporter gene, we spotted the different reporter strains on rich medium, minimal medium lacking uracil and on minimal medium containing 5-FOA (Figure 1C). The 2xSRE reporter strain showed reduced growth on a medium lacking uracil, whereas the other strains grew well. Conversely, strains with four or more copies of the SRE sequence showed reduced growth on a medium containing 5-FOA. All strains grew equally on rich medium (Figure 1C). Reporter gene expression required Sre1N insomuch as deletion of sre1N in the 7xSRE reporter strain blocked growth on a medium lacking uracil, but restored growth to cells plated on a medium containing 5-FOA (Figure 1C, lower panels). Thus, the sre1N SRE-ura4+ reporter strains display Sre1-dependent growth. We conclude that increasing the number of SRE sequences results in increased ura4+ expression and differential growth on uracil- and 5-FOA-containing media.
To identify genes that increase Sre1N transcriptional activity, we screened a cDNA library for plasmids that permitted growth of the sre1N 2xSRE-ura4+ reporter strain on a medium lacking uracil. We chose this strain because insertion of one additional copy of the SRE sequence permitted growth on this medium (Figure 1C), suggesting that growth of this strain would be most sensitive to an increase in Sre1N activity. The plasmid library contained cDNAs expressed from the strong alcohol dehydrogenase promoter (Janoo et al, 2001). We screened 4.56 × 105 transformants for growth on a medium lacking uracil and performed two secondary assays to isolate clones with increased Sre1N transcriptional activity. First, we selected clones that failed to grow on a medium containing 5-FOA. Although the sre1N 2xSRE-ura4+ cells grew on a medium containing 5-FOA, cells with elevated Sre1N activity should not (Figure 1C). Second, Sre1N was assayed by western blotting analysis. Finally, plasmids were recovered from positive clones and retransformed into the sre1N 2xSRE-ura4+ strain to demonstrate that the observed phenotypes were plasmid dependent. From this cDNA library screen, we isolated plasmids coding for four different genes that allowed growth on a medium lacking uracil (Table I). Plasmids coding for SPCC4B3.07+ were recovered most frequently (31/35 plasmids) and this gene was further characterized.
Table 1.
Positive regulators of Sre1N
| Name | Description | Clones isolated from screen | Sre1N level |
|---|---|---|---|
| SPBC1773.16+ | Transcription factor | 1 | ↑↑ |
| SPCC4B3.07+ | Negative regulator of Ofd1 | 31 | ↑↑↑↑ |
| hhp1+ | Casein kinase I family | 2 | ↑ |
| SPBP8B7.08+ | Leucine carboxyl methyltransferase | 1 | — |
| Strain: sre1N 2xSRE-ura4+ cells. | |||
Nro1 is a positive regulator of Sre1N
A plasmid expressing SPCC4B3.07+ allowed growth of the sre1N 2xSRE-ura4+ strain on a medium lacking uracil (Figure 1D). SPCC4B3.07+ codes for a non-essential protein of 393 amino acids that is a putative nuclear pore-associated protein and contains no predicted transmembrane segments (Chen et al, 2004). On the basis of our results below, SPCC4B3.07+ was named nro1+, negative regulator of Ofd1. First, we assayed the ability of nro1+ to regulate Sre1N. sre1N 2xSRE-ura4+ cells carrying empty vector or nro1+ were shifted to low oxygen at increasing times and immunoblotted for Sre1N. As expected, Sre1N increased under low oxygen in cells carrying an empty vector due to decreased Sre1N degradation and the ability of Sre1N to upregulate its own expression through a positive feedback loop (Figure 1E, lanes 1–4) (Hughes and Espenshade, 2008). Cells overexpressing nro1+ had increased Sre1N in the presence of oxygen and elevated Sre1N at each low oxygen time point as compared with cells carrying an empty vector (Figure 1E, lanes 5–8). To examine the requirement of nro1+ for Sre1N expression, we compared sre1N and sre1N nro1Δ cells. Strikingly, although both strains showed equal amounts of Sre1N in the presence of oxygen, deletion of nro1+ dramatically reduced Sre1N in the absence of oxygen (Figure 1F, lanes 5–8), indicating that nro1+ is required for the low oxygen upregulation of Sre1N. Taken together, these data indicate that nro1+ is a positive regulator of Sre1N expression required for the accumulation of Sre1N under low oxygen.
Nro1 regulates Sre1N degradation
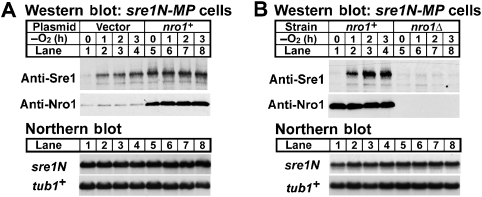
Previous studies demonstrated that sre1+ mRNA is not subject to translational control by oxygen (Sehgal et al, 2008), suggesting that Nro1 regulates Sre1N by controlling either transcription or protein stability. To test whether Nro1 regulates Sre1N through a post-transcriptional mechanism, we examined the function of Nro1 in sre1N-MP (mutant promoter) cells, which lack positive feedback regulation of sre1N transcription due to mutation of the Sre1 DNA-binding sequences in the sre1+ promoter (Hughes and Espenshade, 2008). In sre1N-MP cells, sre1N mRNA does not increase under low oxygen, allowing us to examine post-transcriptional regulation (Figure 2, lower panels). We cultured sre1N-MP cells containing empty vector or overexpressing nro1+ in the presence or absence of oxygen at increasing times. In sre1N-MP cells carrying an empty vector, Sre1N increased upon shifting to low oxygen (Figure 2A, upper panel, lanes 1–4). The increase under low oxygen is reduced as compared with sre1N cells due to the absence of positive feedback regulation at the sre1N promoter in these cells (compare Figure 2A with Figure 1E). Overexpression of nro1+ in sre1N-MP cells increased Sre1N in the presence of oxygen, and cells showed no further increase in Sre1N under low oxygen (Figure 2A, upper panel, lanes 5–8). Conversely, deletion of nro1+ blocked the accumulation of Sre1N under low oxygen in sre1N-MP cells (Figure 2B). Both low oxygen growth and overexpression of nro1+ had no effect on the level of sre1N mRNA (Figure 2, lower panels), indicating that the observed effects of Nro1 on Sre1N were post-transcriptional. In addition, the level of endogenous Nro1 was not affected by oxygen (Figure 2A and B, upper panel, lanes 1–4). Taken together, these results demonstrate that Nro1 is required for the accumulation of Sre1N under low oxygen and that Nro1 regulates Sre1N by a post-transcriptional mechanism.
Figure 2.
Nro1 regulates Sre1N post-transcriptionally. (A) sre1N-MP cells containing empty vector or a plasmid expressing nro1+ from the adh promoter were cultured in a minimal medium without oxygen. Whole-cell extracts (40 μg) were subjected to western blot analysis using anti-Sre1 antibody. Total RNA (10 μg) was subjected to northern blot analysis using sre1N and tub1+ probe for α-tubulin as denoted. (B) sre1N-MP and sre1N-MP nro1Δ cells were cultured in the rich media in the absence of oxygen. Whole-cell extracts and total RNA were analysed as in (A).
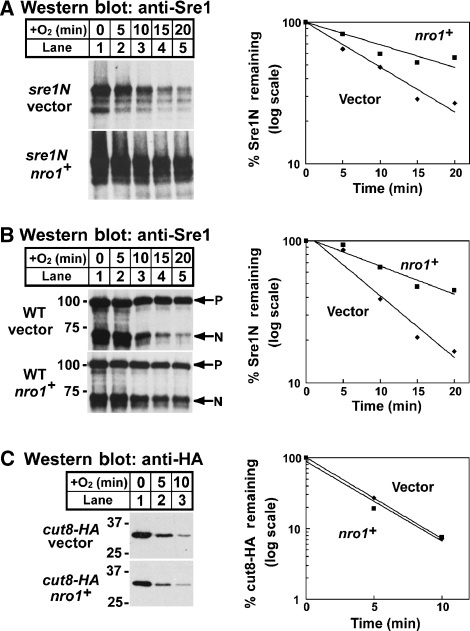
Sre1N is rapidly degraded in the presence of oxygen and Ofd1 accelerates Sre1N degradation (Hughes and Espenshade, 2008). To determine whether Nro1 regulates Sre1N protein stability, we grew sre1N cells containing an empty vector or overexpressing nro1+ in the absence of oxygen for 3 h to accumulate Sre1N. At time t=0, cycloheximide was added to inhibit protein translation, and cells were shifted to the presence of oxygen. The half-life of Sre1N in control cells was 9 min. Overexpression of nro1+ increased the half-life of Sre1N to 19 min, indicating that Nro1 expression stabilizes Sre1N (Figure 3A). Consistent with the results in sre1N cells, we found that overexpression of nro1+ increased the half-life of Sre1N in wild-type cells from 7 to 15 min (Figure 3B). Degradation of another short-lived nuclear protein Cut8-HA was unaffected by nro1+ overexpression (Figure 3C), suggesting that Nro1 does not broadly inhibit protein degradation (Takeda and Yanagida, 2005). Taken together, these data suggest that Nro1 is required for Sre1N accumulation under low oxygen and that Nro1 functions as a positive regulator of Sre1N by controlling Sre1N degradation.
Figure 3.
Nro1 regulates Sre1N degradation. (A) sre1N cells containing empty vector or a plasmid expressing nro1+ were grown in the absence of oxygen for 3 h. At t=0, cycloheximide (200 μg/ml) was added and cells were shifted to the presence of oxygen. (B) Wild-type cells containing empty vector or a plasmid expressing nro1+ were grown in the absence of oxygen for 3 h. At t=0, cycloheximide (200 μg/ml) was added and cells were shifted to the presence of oxygen. P and N denote precursor and N–terminus, respectively. (C) cut8-HA cells containing empty vector or a plasmid expressing nro1+ were grown in the absence of oxygen for 3 h. At t=0, cycloheximide (200 μg/ml) was added and cells were shifted to the presence of oxygen. For each experiment, samples were collected at the indicated times. Whole-cell extracts (40 μg) were subjected to western blot analysis using anti-Sre1 IgG or anti-HA IgG as indicated. The percentage of Sre1N or Cut8-HA remaining at each time point relative to t=0 is quantified to the right; vector (diamonds) and nro1+ (squares). Data shown are representative of four experiments.
Nro1 is a negative regulator of Ofd1
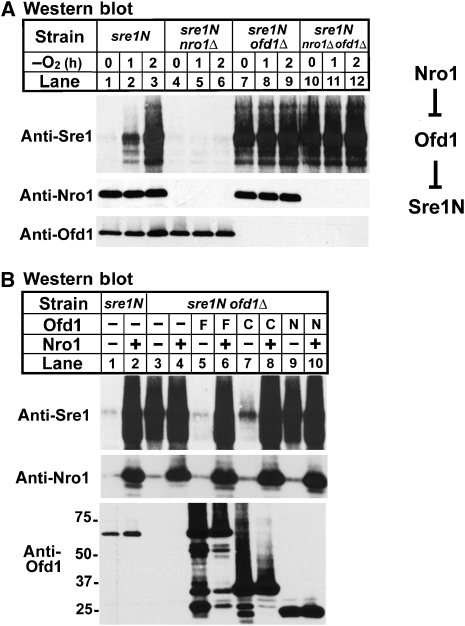
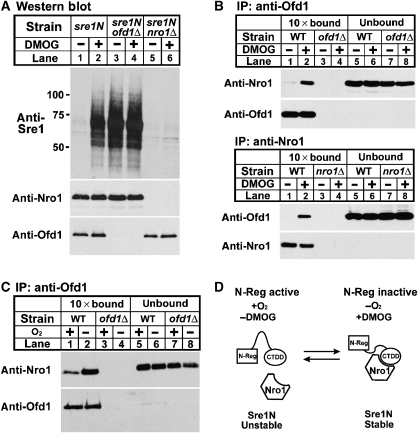
In the presence of oxygen, Ofd1 accelerates the degradation of Sre1N, and inhibition of Ofd1 under low oxygen stabilizes Sre1N (Hughes and Espenshade, 2008). Interestingly, we noted that sre1N-MP cells overexpressing nro1+ resembled sre1N-MP ofd1Δ cells. Both strains have elevated levels of Sre1N in the presence of oxygen and loss of oxygen regulation, suggesting that Nro1 stabilizes Sre1N by acting through Ofd1 (Figure 2A) (Hughes and Espenshade, 2008). To test this, we assayed the genetic interaction between nro1+ and ofd1+. We cultured sre1N, sre1N nro1Δ, sre1N ofd1Δ and sre1N nro1Δ ofd1Δ cells in the presence or absence of oxygen and assayed Sre1N by western blotting. Under low oxygen, Sre1N accumulated in sre1N cells (Figure 4A, lanes 1–3). As expected, deleting nro1+ or ofd1+ had opposite effects on the level of Sre1N (Figure 4A, lanes 4–9). In nro1Δ cells, Sre1N failed to accumulate under low oxygen, and in ofd1Δ cells Sre1N accumulated in the presence of oxygen and showed no further increase under low oxygen. Importantly, sre1N nro1Δ ofd1Δ cells were identical to sre1N ofd1Δ cells, having elevated Sre1N in the presence of oxygen and no further increase under low oxygen (Figure 4A, lanes 7–12). Therefore, deleting ofd1+ from sre1N nro1Δ cells increased Sre1N, indicating that the ability of Nro1 to regulate Sre1N requires Ofd1. From these data, we conclude that (1) Nro1 functions upstream of Ofd1 and (2) Nro1 functions as a positive regulator of Sre1N by inhibiting the negative regulator Ofd1. The genetic relationship between Nro1 and Ofd1 in the regulation of Sre1N turnover is shown on the right of Figure 4A.
Figure 4.
Nro1 inhibits Ofd1-dependent degradation of Sre1N. (A) sre1N, sre1N nro1Δ, sre1N ofd1Δ and sre1N nro1Δ ofd1Δ cells were grown in the absence of oxygen for the indicated times. Whole-cell extracts (40 μg) were subjected to western blot analysis using anti-Sre1, anti-Nro1 or anti-Ofd1 antibody as indicated. Genetic interactions among genes are shown on the right. (B) sre1N and sre1N ofd1Δ cells containing empty vector, expressing full-length Ofd1 (F), the Ofd1 C terminus (C) or the Ofd1 N terminus (N) and also containing either empty vector or expressing nro1+ were cultured in a minimal medium for 20 h. Whole-cell extracts (40 μg) were subjected to western blot analysis using anti-Sre1, anti-Nro1 or anti-Ofd1 antibody as indicated.
To further test whether Nro1 regulates Sre1N by inhibiting Ofd1, we generated sre1N ofd1Δ cells overexpressing full-length Ofd1, the Ofd1 C terminus (aa 242–515) or the Ofd1 N terminus (aa 1–241), with or without overexpression of Nro1, and we assayed Sre1N accumulation. The Ofd1 C terminus has been shown to be sufficient to accelerate Sre1N degradation and is referred to as the CTDD (Hughes and Espenshade, 2008). Sre1N was stabilized in sre1N cells overexpressing Nro1 (Figure 4B, lanes 1 and 2). Deletion of ofd1+ stabilizes Sre1N, and expression of either full-length Ofd1 or the Ofd1 CTDD, but not the Ofd1 N terminus, accelerated Sre1N degradation (Figure 4B, lanes 3, 5, 7 and 9) (Hughes and Espenshade, 2008). Importantly, overexpression of Nro1 blocked the ability of full-length Ofd1 and the Ofd1 CTDD to degrade Sre1N (Figure 4B, lanes 6 and 8). Given that Nro1 stabilizes Sre1N (Figure 3), these results suggest that Nro1 functions by inhibiting the ability of the Ofd1 CTDD to accelerate Sre1N degradation.
Nro1 binds to the Ofd1 CTDD
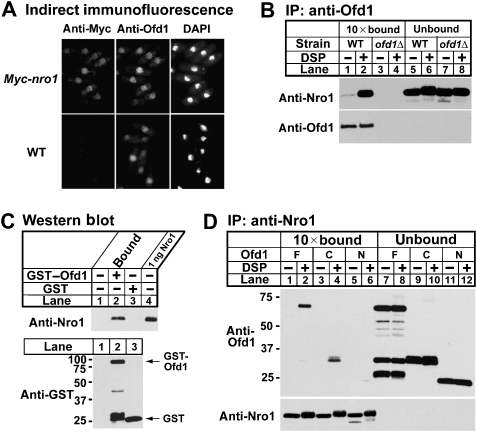
To determine the intracellular localization of endogenous Nro1, we performed indirect immunofluorescence using a chromosomal (His)6Myc-tagged nro1 strain. (His)6Myc-nro1 cells showed oxygen-dependent regulation of Sre1N degradation, indicating that (His)6Myc-Nro1 was functional (data not shown). Nro1 was enriched in the nucleus with some cytosolic staining (Figure 5A). Staining for Nro1 was specific as wild-type cells expressing untagged Nro1 showed no signal. The Saccharomyces cerevisiae homologue of Nro1, YOR051C, has been reported to be associated with nuclear pore complexes (Rout et al, 2000). Our confocal microscopy studies indicated that Nro1 was concentrated in the nucleus, but not in the nuclear rim (data not shown). This result was consistent with recent global protein localization studies indicating that an Nro1–YFP fusion protein localized primarily to the nucleus (Matsuyama et al, 2006). To determine whether Nro1 colocalizes with Ofd1, endogenous Ofd1 was visualized using an affinity-purified anti-Ofd1 antibody. Ofd1 colocalized with Nro1 in the nucleus, demonstrating that Nro1 and Ofd1 localize to the same subcellular compartment (Figure 5A).
Figure 5.
Nro1 binds the C terminus of Ofd1. (A) (His)6Myc-nro1 and wild-type cells were analysed by indirect immunofluorescence using anti-Myc antibody, anti-Ofd1 antibody and 4′,6-diamidino-2-phenylindole (DAPI) to stain DNA. (B) Wild-type and ofd1Δ cells grown in rich medium were treated with either 2% DMSO or 2 mM DSP crosslinker in PBS for 5 min. Detergent-solubilized whole-cell extracts were subjected to immunoprecipitation with anti-Ofd1. Bound (10-fold overloaded) and unbound fractions were analysed by western blot analysis with anti-Nro1–HRP and anti-Ofd1–HRP antibodies. (C) Purified recombinant 6 × His–Nro1 was incubated at room temperature for 45 min with glutathione-agarose beads preloaded with GST–Ofd1, GST or buffer only. Bound fractions were analysed by western blotting with anti-Nro1 and anti-GST antibodies. 1 ng of Nro1 was loaded in lane 4. (D) sre1N ofd1Δ cells overexpressing full-length Ofd1, the Ofd1 C terminus or the Ofd1 N terminus from the thiamine-repressible nmt promoter were cultured in a minimal medium for 20 h. Cells were treated with either 2% DMSO or 2 mM DSP crosslinker in PBS for 5 min. Detergent-solubilized whole-cell extracts were subjected to immunoprecipitation with anti-Nro1. Bound (10-fold overloaded) and unbound fractions were analysed by western blot analysis with anti-Ofd1–HRP and anti-Nro1–HRP antibodies.
On the basis of the observation that Nro1 is an inhibitor of the Ofd1 CTDD, we next asked whether Nro1 forms a complex with Ofd1 in a co-immunoprecipitation experiment. Using anti-Ofd1 antibody, Nro1 copurified with endogenous Ofd1 in extracts from wild type, but not ofd1Δ cells. Binding between Ofd1 and Nro1 increased upon addition of the reversible crosslinker dithiobis-succinimidyl propionate (DSP) (Figure 5B, lanes 1–4). To test whether Nro1 binds Ofd1 directly, purified recombinant Nro1 was incubated with glutathione-agarose beads preloaded with purified GST–Ofd1 or GST. Nro1 bound to GST–Ofd1 beads, but not GST beads, demonstrating that Nro1 binds directly to Ofd1 (Figure 5C, lanes 2 and 3).
To map where Nro1 binds Ofd1, we tested binding between Nro1 and different fragments of Ofd1. We immunoprecipitated Nro1 from sre1N ofd1Δ cells expressing full-length Ofd1, the Ofd1 CTDD (aa 242–515) or the Ofd1 N terminus (aa 1–241), and bound fractions were probed for Ofd1. Endogenous Nro1 formed a complex with full-length Ofd1 and the Ofd1 CTDD, but not the Ofd1 N terminus (Figure 5D, lanes 1–6). Given that Nro1 also inhibits the ability of the Ofd1 CTDD to degrade Sre1N (Figure 4B), these data suggest that Nro1 regulates Sre1N turnover by directly binding to the Ofd1 CTDD.
Ofd1 iron-coordinating mutants that accelerate Sre1N degradation display reduced Nro1 binding
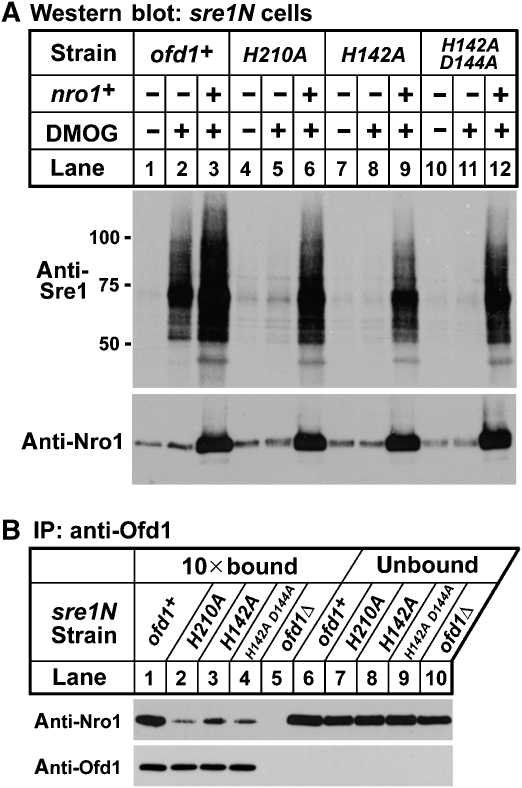
The N terminus of Ofd1 contains a prolyl 4-hydroxylase-like 2-OG–Fe(II) dioxygenase domain, which is an iron- and oxygen-dependent enzyme inhibited under low oxygen and by the chemical dimethyloxalylglycine (DMOG) (Ozer and Bruick, 2007). The dioxygenase domain of Ofd1 regulates the ability of the CTDD to accelerate the degradation of Sre1N (Hughes and Espenshade, 2008). In the presence of oxygen when the dioxygenase is predicted to be active, Ofd1 accelerates Sre1N degradation, but under low oxygen or in the presence of DMOG Sre1N is stabilized. Inhibition of Ofd1 in sre1N cells by treatment with DMOG increased Sre1N (Figure 6A, lanes 1 and 2). Previous studies showed that mutation of the iron-coordinating residues in the dioxygenase domain made Ofd1 unresponsive to DMOG, but unexpectedly the iron-coordinating mutants were constitutively active for Sre1N degradation (Figure 6A, lanes 4, 5, 7, 8, 10 and 11) (Hughes and Espenshade, 2008). To investigate whether the constitutive activity of Ofd1 mutants results from a change in Nro1 binding, we examined the interaction of the Ofd1 iron-coordinating mutants and Nro1 by co-immunoprecipitation (Figure 6B). Wild-type Ofd1 efficiently formed a complex with Nro1 (Figure 6B, lane 1). Interestingly, each of the Ofd1 iron-coordinating mutants showed reduced binding to Nro1 (Figure 6B, lanes 2–4), despite equal expression of the Ofd1 mutants. These data suggest that the altered activity of the Ofd1 mutants results from reduced binding to Nro1.
Figure 6.
Constitutively active Ofd1 mutants are defective for binding to Nro1. (A) sre1N, sre1N ofd1-H210A, sre1N ofd1-H142A and sre1N ofd1-H142A D144A cells containing empty vector or nro1+ plasmid were treated with either 1% DMSO or 20 mM DMOG for 3 h. Whole-cell extracts (40 μg) were subjected to western blot analysis using anti-Sre1 and anti-Nro1 antibodies. (B) sre1N, sre1N ofd1-H210A, sre1N ofd1-H142A, sre1N ofd1-H142A D144A and sre1N ofd1Δ cells cultured in rich medium were treated with 2 mM DSP crosslinker in PBS for 5 min. Detergent-solubilized whole-cell extracts were subjected to immunoprecipitation with anti-Ofd1. Bound (10-fold overloaded) and unbound fractions were analysed by western blot with anti-Nro1–HRP and anti-Ofd1–HRP antibodies.
To determine whether Nro1 could inhibit the constitutively active Ofd1 mutants, we expressed Nro1 from a plasmid in wild-type and Ofd1 mutant cells in the presence of oxygen. Despite the increased activity of the Ofd1 mutants, overexpression of Nro1 suppressed the constitutively active Ofd1 mutants, resulting in the accumulation of Sre1N (Figure 6A, lanes 6, 9 and 12). Collectively, these results suggest that Nro1 binding to Ofd1 inhibits the ability of Ofd1 to accelerate Sre1N degradation.
N-Reg dioxygenase domain of Ofd1 regulates binding to Nro1
The N-terminal dioxygenase domain of Ofd1 (N-Reg) is required for oxygen sensing and regulation of the CTDD. Previously, we proposed the following model (Hughes and Espenshade, 2008). In the absence of dioxygenase activity due to low oxygen or DMOG treatment, the N-Reg domain inhibits the ability of the CTDD to accelerate Sre1N degradation leading to increased Sre1N. Conversely in the presence of oxygen, active N-Reg dioxygenase no longer inhibits the CTDD, causing rapid degradation of Sre1N (Hughes and Espenshade, 2008). We next tested whether Nro1 is required for N-Reg inhibition of the Ofd1 CTDD using the dioxygenase inhibitor DMOG. We grew sre1N, sre1N ofd1Δ and sre1N nro1Δ cells in the absence or presence of DMOG for 3 h. Consistent with the model, chemical inhibition of the N-Reg dioxygenase domain by DMOG in sre1N cells resulted in the accumulation of Sre1N, and deletion of ofd1+ led to constitutively high Sre1N (Figure 7A, lanes 1–4). Interestingly, in sre1N nro1Δ cells Sre1N did not accumulate when the N-Reg dioxygenase was inhibited by DMOG (Figure 7A, lanes 5 and 6). Thus, Nro1 is required for Sre1N accumulation in DMOG-treated cells, indicating that Nro1 is required for inhibition of the Ofd1 CTDD by the N-Reg dioxygenase domain.
Figure 7.
Oxygen regulates Nro1 binding to Ofd1. (A) sre1N, sre1N ofd1Δ and sre1N nro1Δ cells were treated with either 1% DMSO or 20 mM DMOG for 3 h. Whole-cell extracts (40 μg) were subjected to western blot analysis using the indicated antibodies. (B) Wild-type, ofd1Δ and nro1Δ cells were grown in rich medium and treated with either 1% DMSO or 20 mM DMOG for 3 h with no DSP added. Detergent-solubilized whole-cell extracts were subjected to immunoprecipitation with anti-Ofd1 (upper panels) or anti-Nro1 (lower panels) antibody. Bound (10-fold overloaded) and unbound fractions were analysed by western blot analysis with anti-Nro1–HRP and anti-Ofd1–HRP antibodies. (C) Wild-type and ofd1Δ cells were grown with or without oxygen for 3 h and were treated with 2 mM DSP crosslinker in PBS for 5 min. Samples were processed as in (B). (D) Model for oxygen-regulated binding between Ofd1 and Nro1. Activity of the Ofd1 N-terminal dioxygenase domain (N-Reg) controls binding between Ofd1 and Nro1. In the presence of oxygen, the N-Reg domain of Ofd1 prevents binding of Nro1 to the CTDD, leading to degradation of Sre1N. Under low oxygen or in the presence of the inhibitor DMOG, N-Reg is inactive, allowing Nro1 to bind the Ofd1 CTDD and block Sre1N degradation.
Given that Nro1 inhibits the ability of the Ofd1 CTDD to degrade Sre1N and Nro1 binds Ofd1, we next tested whether binding between Nro1 and Ofd1 was regulated by the activity of N-Reg dioxygenase domain using the inhibitor DMOG. Wild-type and ofd1Δ cells were grown in the presence or absence of DMOG for 3 h and binding between endogenous Nro1 and Ofd1 was assayed by co-immunoprecipitation. Using anti-Ofd1 antibody, a small amount of Nro1 was detected in the bound fraction in untreated cells (Figure 7B, lane 1, upper panels). In the presence of DMOG, Ofd1 binding to Nro1 increased dramatically (Figure 7B, lane 2, upper panels). Binding was specific as no Nro1 was detected in the bound fractions using ofd1Δ cells (Figure 7B, lanes 3 and 4). Ofd1 binding to Nro1 also increased upon DMOG treatment using anti-Nro1 antibody in a reciprocal co-immunoprecipitation experiment (Figure 7B, lanes 1–4, lower panels). Taken together, these data demonstrate that the Ofd1 N-Reg dioxygenase domain regulates binding of Nro1 to Ofd1 and that Nro1 is required for the ability of the N-Reg domain to inhibit the Ofd1 CTDD.
Oxygen regulates binding of Nro1 to Ofd1
Under physiological conditions, Ofd1 regulates Sre1N in response to oxygen availability. To test whether oxygen regulates Nro1 binding to Ofd1, wild-type and ofd1Δ cells were cultured in the presence or absence of oxygen and Nro1–Ofd1 binding was assayed by co-immunoprecipitation. Nro1 bound to Ofd1 in the presence of oxygen, and Nro1 binding to Ofd1 increased significantly under low oxygen (Figure 7C, lanes 1 and 2). These results demonstrate that oxygen regulates binding between Ofd1 and its inhibitor Nro1. Collectively, the experiments in Figure 7 suggest that inhibition of Ofd1 N-Reg dioxygenase activity by DMOG or under low oxygen stimulates binding of Nro1 to Ofd1, resulting in the inhibition of Sre1N degradation by the Ofd1 CTDD.
Discussion
Sre1 is a membrane-bound transcription factor that is a principal regulator of the hypoxic transcriptional response in fission yeast. The active transcription factor Sre1N is released from the membrane by proteolytic cleavage under low oxygen (Hughes et al, 2005). Previously, we demonstrated that the prolyl 4-hydroxylase-like 2-OG–Fe(II) dioxygenase Ofd1 controls the oxygen-regulated degradation of Sre1N (Hughes and Espenshade, 2008). In the present study, we used a genetic screen to identify Nro1 as an inhibitor of Ofd1 and describe the mechanism by which oxygen controls the activity of Ofd1 and Sre1N stability. Our model for this regulatory mechanism is outlined in Figure 7D. In the presence of oxygen, the N-Reg dioxygenase is active and Nro1 is unable to bind and inhibit the Ofd1 CTDD. Consequently, the Ofd1 CTDD accelerates Sre1N degradation and downregulates Sre1N. In the absence of oxygen, the N-Reg dioxygenase is inactive and Nro1 binds to the Ofd1 CTDD, thereby inhibiting the Ofd1 CTDD and stabilizing Sre1N. In this mechanism, the N-Reg dioxygenase functions as an oxygen sensor to regulate binding of the inhibitor Nro1 to the Ofd1 CTDD and to control Sre1N stability.
Multiple independent lines of evidence support this model. First, Nro1 is a positive regulator of Sre1N stability that functions through Ofd1 (Figures 1, 3 and 4A). Second, Nro1 is required for upregulation of Sre1N both under low oxygen and by DMOG treatment (Figures 1F, 2B and 7A). Third, Nro1 inhibits the Ofd1 CTDD and directly binds to Ofd1 in vitro (Figures 4B and 5C). Fourth, Nro1 binds to Ofd1 when N-Reg dioxygenase activity is inhibited under low oxygen or by DMOG (Figure 7B and C). Fifth, Ofd1 iron-coordinating mutants constitutively active for Sre1N degradation are defective for Nro1–Ofd1 binding (Figure 6). Collectively, these data argue that the N-Reg dioxygenase domain controls the oxygen-dependent binding of Nro1 to the Ofd1 CTDD, thereby regulating the stability of Sre1N (Figure 7D).
This oxygen-dependent regulatory mechanism is most likely conserved in S. cerevisiae. Binding between the budding yeast Nro1 (YOR051C) and the Ofd1 homologue Tpa1 has been detected by global affinity-purification studies (Krogan et al, 2006). Although a clear Nro1 homologue is not readily identifiable in mammals, the CTDD of Ofd1 shows significant identity to the C-terminal domain of its human homologue OGFOD1 (Hughes and Espenshade, 2008), suggesting that a similar oxygen-dependent regulatory mechanism exists in humans.
nro1+ was identified by a genetic screen for genes that increase Sre1N transcriptional activity when expressed from a plasmid. This initial screen identified 35 independent clones, 31 for nro1+, 2 for hhp1+ and 1 each for SPBC1771.16+ and SPBP8B7.08+ (Table I). Secondary assays indicate that both hhp1+, a casein kinase I family member, and SPBC1773.16+, a predicted transcription factor, increase Sre1N levels. Given that Sre1N is highly phosphorylated (Hughes et al, 2005), Hhp1 may function directly or indirectly to regulate Sre1N stability or transcriptional activity by phosphorylation. SPBP8B7.08+ expression had no effect on Sre1N levels and encodes a putative leucine carboxyl methyltransferase. Future studies will investigate the regulatory mechanisms of these genes.
The N-terminal 2-OG-Fe(II) dioxygenase domain of Ofd1 belongs to the prolyl 4-hydroxylase enzyme family, which includes the PHD prolyl hydroxylases that control the stability of the mammalian HIF-α hypoxic transcription factors (Schofield and Ratcliffe, 2005; Gordan and Simon, 2007). Proline hydroxylation of HIF-α results in its recognition and ubiquitination by the von Hippel–Lindau protein-containing E3 ligase and subsequent proteasomal degradation (Kaelin, 2005). In contrast, our studies demonstrate that Ofd1 accelerates Sre1N degradation by a mechanism that requires the CTDD, but not the dioxygenase domain (Hughes and Espenshade, 2008). How the Ofd1 CTDD facilitates Sre1N turnover is unknown. In addition, our data indicate that the activity of the N-Reg dioxygenase controls Nro1 binding and the function of the Ofd1 CTDD. It remains to be determined whether the substrate for this dioxygenase domain is another protein or perhaps a small molecule. Current studies focus on these unanswered questions.
Materials and methods
Materials
Yeast extract was obtained from Becton Dickinson and Co.; amino acids for the medium from Sigma; Edinburgh minimal medium from MP Biomedical; horseradish peroxidase-conjugated affinity-purified donkey anti-rabbit and anti-mouse immunoglobulin G (IgG) from Jackson ImmunoResearch; anti-HA 12CA5 mouse monoclonal antibody from Roche; oligonucleotides from Integrated DNA Technologies; DMOG from Frontier Scientific Inc. and DSP crosslinker from Pierce.
Yeast strains and culture
Wild-type haploid S. pombe KGY425 (h-, his3-D1, leu1-32, ura4-D18 and ade6-M210) was obtained from American Type Culture Collection (Burke and Gould, 1994). S. pombe strain sre1Δ has been described earlier (Hughes et al, 2005). S. pombe strains sre1N, sre1N-MP, sre1N ofd1Δ and cut8-3xHA, and strains containing mutations in Ofd1 iron-coordinating residues at the endogenous ofd1+ locus have been described earlier (Hughes and Espenshade, 2008). S. pombe nro1Δ, ofd1Δ and nro1Δ ofd1Δ were generated from haploid yeast (KGY425, KGY461, sre1N or sre1N-MP) by homologous recombination using established techniques (Bahler et al, 1998; Hughes and Espenshade, 2008). Strain (His)6Myc-nro1 contains N-terminal (His)6Myc tag that was generated by the Cre-loxP method (Werler et al, 2003). The sre1N SRE-ura4+ reporter strains were generated by crossing SRE-ura4+-kanMX (Hughes et al, 2008) with sre1N::kanMX. The sre1N::kanMX strain contains a precise deletion of the sequence coding for Sre1 amino acids 441–900 and was generated in KGY425 background by homologous recombination (Bahler et al, 1998).
Yeast strains were grown to exponential phase at 30°C in yeast extract plus supplements (225 μg/ml each of histidine, leucine, adenine, lysine and uracil) or in Edinburgh minimal medium where indicated using standard techniques (Hughes et al, 2005). Anaerobic growth conditions were maintained using an In vivo2 400 workstation (Biotrace Inc.) as described earlier (Hughes et al, 2005; Todd et al, 2006). Minimal medium containing 0.1% 5-FOA (Toronto Research Chemicals) was made using a standard recipe (Boeke et al, 1987; Moreno et al, 1991).
Plasmids
Ofd1 constructs have been described earlier (Hughes and Espenshade, 2008). Plasmid expressing nro1+ cDNA from the adh promoter was isolated from the screen for positive regulators. Plasmids containing nro1+ expressed from the thiamine-repressible nmt promoter were generated by the insertion of nro1+ PCR fragments into XhoI–SmaI sites of pREP4X (Forsburg, 1993).
Plasmid cDNA library screen
sre1N 2xSRE-ura4+ cells transformed with the SPLE-1 S. pombe cDNA library were plated on a minimal medium lacking leucine and uracil (Okazaki et al, 1990; Janoo et al, 2001). A total of 4.56 × 105 transformants were screened for growth, and transformants that grew up within 5–8 days were picked for single colony purification. Transformants were screened for growth on a medium containing 5-FOA. For clones that failed to grow on 5-FOA, plasmid DNA was extracted, amplified in Escherichia coli and sequenced using the oligonucleotide adh-forward 5′-TCTCATTGGTCTTCCGCTCCG-3′. cDNA expression was driven by the adh promotor in a vector pLEV3 containing S. cerevisiae LEU2 as a selectable marker. Library SPLE-1 contains ∼6 × 105 clones, 67% of which carry inserts >1 kb (Janoo et al, 2001).
Antibody preparation
Polyclonal antiserum against full-length Nro1 was generated by immunizing rabbits with bacterially expressed antigen using a standard protocol (Covance). A bacterial expression plasmid for Nro1 was created by cloning PCR products generated using nro1+ cDNA template isolated from genetic screen into the expression vector pPROEX HTb (Gibco BRL). Antigen containing an N-terminal 6 × His fusion to Nro1 was purified from E. coli using Ni-NTA agarose (Qiagen). Affinity-purified anti-Nro1 antibody (anti-Nro1 IgG) was isolated from rabbit serum by affinity chromatography with a resin coupled to Nro1 antigen using the AminoLink Plus Immobilization Kit (Pierce) according to the manufacturer's instructions. Affinity-purified anti-Nro1–HRP and anti-Ofd1–HRP were generated using EZ-Link Plus Activated Peroxidase kit (Pierce).
Indirect immunofluorescence, northern and western blotting
Indirect immunofluorescence staining was performed as described earlier using anti-myc antibody (1 μg/ml) (Santa Cruz Biotechnology) and 20 μg/ml goat anti-mouse Alexa 488 antibody (Molecular Probes) (Hagan and Hyams, 1988; Hughes and Espenshade, 2008). Cell images were taken using the IVISION-Mac V.4.07 software. All images were normalized to control cells to minimize nonspecific signal and then processed uniformly using Adobe Photoshop to optimize the image contrast. Total RNA isolation and northern blot analysis were performed as described earlier (Hughes et al, 2005). PCR fragments for sre1N and tub1+ used for northern probe synthesis were generated using gene-specific primer pairs (Supplementary Table 1). Whole-cell yeast extract preparation and western blot analysis using anti-Sre1 IgG and horseradish peroxidase-conjugated anti-rabbit IgG were performed as described earlier (Hughes and Espenshade, 2008). Anti-Nro1 serum was used at 1:10 000 dilution.
GST pull-down assay
A bacterial expression plasmid for an N-terminal GST fusion to Ofd1 (GST–Ofd1) was created by cloning a PCR product generated using S. pombe genomic DNA as a template into the expression vector pGEX-KG (Guan and Dixon, 1991). GST–Ofd1 and GST were purified from E. coli using glutathione-agarose (Sigma). An N-terminal 6 × His fusion to Nro1 was purified from E. coli according to the manufacturer's instructions (Qiagen). 6 × His–Nro1 (108 μM=∼2.5 μg) was incubated with glutathione-agarose preloaded with GST or GST–Ofd1 protein (33 μM) in 1 ml RIPA buffer (150 mM NaCl, 0.5% sodium deoxycholate, 50 mM Tris–HCl pH 7.4 and 1% NP-40) containing 2 μg BSA and protease inhibitors for 45 min at room temperature. Agarose beads were washed with RIPA buffer three times. Bound fractions were analysed by SDS–PAGE and western blotting using anti-Nro1 and anti-GST (Covance; MMS-112R) antibodies.
Measurement of protein turnover
For protein half-life analysis, yeast strains were grown in the absence of oxygen for 3 h to exponential phase and then shifted to normoxic conditions in the presence of cycloheximide (200 μg/ml) (Sigma). Samples were harvested, and whole-cell lysates (40 μg) were subjected to western blot analysis. Blots were developed using HyGLO HRP Detection kit (Denville Scientific). Blots were imaged using the Versadoc Imaging System (Bio-Rad), and signals were quantified by using Quantity One Software (Bio-Rad). An exponential trend line was added and used to calculate the line slope (k). Protein half-life was calculated using the equation: T1/2=(ln 2)/k (Belle et al, 2006).
Co-immunoprecipitation
For Nro1 and Ofd1 co-immunoprecipitation, cells (5 × 107) were lysed using glass beads (0.5 mm; Sigma) in 100 μl NP-40 lysis buffer (50 mM Hepes, pH 7.4, 100 mM NaCl, 1.5 mM MgCl2 and 1%(v/v) NP-40) plus protease inhibitors (Hughes et al, 2005). Insoluble material was removed by centrifugation at 2 × 104 g for 2 min, and the supernatant was subjected to immunoprecipitation in 1 ml NP-40 lysis buffer for 2 h using 5 μg of affinity-purified anti-Ofd1 IgG or anti-Nro1 IgG and 30 μl protein A beads (Repligen). Beads were washed three times with NP-40 lysis buffer, and co-immunopurified proteins were eluted by boiling in SDS–PAGE loading buffer (Hughes et al, 2005).
Supplementary Material
Supplementary Table 1
Acknowledgments
We thank Espenshade Lab members for experimental suggestions and for reviewing the paper. We kindly thank Charlie Hoffman (Boston College University) for providing the S. pombe cDNA plasmid library and David Balasundaram (IMCB, A-STAR, Singapore) for an nro1Δ strain used in preliminary experiments. This study was supported by a grant from the National Institutes of Health (HL-077588). C-YSL and BTH are recipients of American Heart Association Predoctoral Fellowships, and PJE is an Established Investigator of the American Heart Association.
References
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK (2006) Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA 103: 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Burke JD, Gould KL (1994) Molecular cloning and characterization of the Schizosaccharomyces pombe his3 gene for use as a selectable marker. Mol Gen Genet 242: 169–176 [DOI] [PubMed] [Google Scholar]
- Chen XQ, Du X, Liu J, Balasubramanian MK, Balasundaram D (2004) Identification of genes encoding putative nucleoporins and transport factors in the fission yeast Schizosaccharomyces pombe: a deletion analysis. Yeast 21: 495–509 [DOI] [PubMed] [Google Scholar]
- Dann CE III, Bruick RK (2005) Dioxygenases as O2-dependent regulators of the hypoxic response pathway. Biochem Biophys Res Commun 338: 639–647 [DOI] [PubMed] [Google Scholar]
- Emerling BM, Chandel NS (2005) Oxygen sensing: getting pumped by sterols. Sci STKE 2005: e30. [DOI] [PubMed] [Google Scholar]
- Espenshade PJ, Hughes AL (2007) Regulation of sterol synthesis in eukaryotes. Annu Rev Genet 41: 401–427 [DOI] [PubMed] [Google Scholar]
- Forsburg SL (1993) Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res 21: 2955–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS (2006) Protein sensors for membrane sterols. Cell 124: 35–46 [DOI] [PubMed] [Google Scholar]
- Gordan JD, Simon MC (2007) Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev 17: 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Dixon JE (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem 192: 262–267 [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS (1988) The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci 89: 343–357 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Lee CS, Bien CM, Espenshade PJ (2007) 4-Methyl sterols regulate fission yeast SREBP-Scap under low oxygen and cell stress. J Biol Chem 282: 24388–24396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Stewart EV, Espenshade PJ (2008) Identification of twenty-three mutations in fission yeast Scap that constitutively activate SREBP. J Lipid Res 49: 2001–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Todd BL, Espenshade PJ (2005) SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120: 831–842 [DOI] [PubMed] [Google Scholar]
- Hughes BT, Espenshade PJ (2008) Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J 27: 1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoo RT, Neely LA, Braun BR, Whitehall SK, Hoffman CS (2001) Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors and the CCAAT binding factor activation complex. Genetics 157: 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG (2005) Proline hydroxylation and gene expression. Annu Rev Biochem 74: 115–128 [DOI] [PubMed] [Google Scholar]
- Krogan NJ et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Kwast KE, Burke PV, Poyton RO (1998) Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J Exp Biol 201: 1177–1195 [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, Horinouchi S, Yoshida M (2006) ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24: 841–847 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H (1990) High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res 18: 6485–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer A, Bruick RK (2007) Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol 3: 144–153 [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148: 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield CJ, Ratcliffe PJ (2005) Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun 338: 617–626 [DOI] [PubMed] [Google Scholar]
- Sehgal A, Hughes BT, Espenshade PJ (2008) Oxygen-dependent, alternative promoter controls translation of tco1+ in fission yeast. Nucleic Acids Res 36: 2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Lee CY, Espenshade PJ (2007) SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet 3: 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Yanagida M (2005) Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell 122: 393–405 [DOI] [PubMed] [Google Scholar]
- Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ (2006) Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol 26: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werler PJ, Hartsuiker E, Carr AM (2003) A simple Cre-loxP method for chromosomal N-terminal tagging of essential and non-essential Schizosaccharomyces pombe genes. Gene 304: 133–141 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1