EMBO J 28, 123–134 (2009); published online 21 January 2009
A thought-provoking study in this issue of The EMBO Journal shows that the circadian clock in mouse fibroblasts is surprisingly insensitive to the inhibition of total cellular mRNA production. The authors go on to show intriguing parallels between compensation of period to changes in temperature and global transcription rate.
The continual progression between day and night has applied a constant selective pressure upon organisms to anticipate, and entrain to, periodic changes in their environment. Cells and organisms have thus evolved endogenous daily timekeeping mechanisms that facilitate the temporal organisation of metabolic, physiological and behavioural processes, for example, the sleep/wake cycle (Gachon et al, 2004). These circadian (meaning ‘about daily') rhythms are observed in all eukaryotes, with as much as 20% of cellular gene expression being under circadian control in mammals (Reddy et al, 2006).
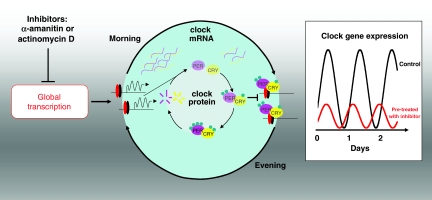
The molecular basis of cellular rhythmicity has been described as comprising several interlinked transcriptional/translational feedback loops whereby clock genes, directly or indirectly, repress their own transcription (Figure 1). In mammals, rhythmically expressed clock genes, such as Period (PER) (1 and 2), Cryptochrome (CRY) (1 and 2) and Bmal1, are essential for accurate time-keeping, as mice lacking them exhibit altered period, or are arrhythmic, at both the behavioural and cellular level (Liu et al, 2007). Therefore, in current models, the timing and amplitude of clock gene expression is critical to function (Forger and Peskin, 2003).
Figure 1.
Schematic showing the effect of transcriptional inhibition on the core transcriptional/translational feedback loop of the mammalian circadian clock. Clock and Bmal1 transcription factors (red/black ovals) bind to E-box promoter elements, activating transcription of Period (PER) and Cryptochrome (CRY) genes around subjective dawn. Following translation, PER/CRY are modified post-translationally (e.g., by phosphorylation—green circles) before nuclear import, wherein they repress their own transcription. These inhibitory complexes are broken down during subjective night. Ancillary loops (not shown) produce rhythmic expression of Bmal1. Pre-treatment of cells with transcriptional inhibitors reduces global transcription rates, and has the effect of reducing the amplitude and period of clock gene expression (box).
Unlike most enzymatic reactions, which approximately double in rate for each 10°-temperature increase (Q10∼2), circadian rhythms are temperature compensated over the biological range (Q10∼0.8–1.4). For example, fibroblasts cultured in vitro display a 26-h period at 37°C, but a 24-h period at 31°C (Dibner et al, 2009). As many cellular properties change markedly with temperature, the basis for this aspect of the cellular oscillator's robustness has remained elusive (Akman et al, 2008).
Recently, several groups have reported observations that challenge the central hypothesis that fine temporal control of transcription levels lies at the heart of circadian timekeeping. For example, constitutive over-expression of auto-repressors CRY1/2 does not affect rhythmicity (Fan et al, 2007), whereas perturbations that affect cyclic AMP signalling do so, dramatically (O'Neill et al, 2008). Now, Dibner et al (2009) have shown clearly that the cellular oscillator is remarkably resilient to large-scale changes in global transcription. In this study, mouse fibroblasts were pre-treated with well-known transcriptional inhibitors α-amanitin and actinomycin D. Following treatment, a reduction of >70% in total mRNA production was observed, and RT–PCR and western blots confirmed that clock gene expression was similarly affected. According to most of the circadian models tested, this should have resulted in arrhythmicity or lengthened period, as indeed do several other pharmacological effectors of the clock (Hastings et al, 2008).
Intriguingly, not only did the cultures in which transcription was reduced remain rhythmic, albeit with lower amplitude, but also inhibitor-treated cells ran significantly faster, by 2–3 h at 37°C. This effect was evident both at the whole culture and at the single-cell level.
Most surprisingly, the period of inhibitor-treated cultures was not as sensitive to changes in temperature as were control cultures (Q10 closer to 1). Furthermore, fibroblasts lacking the clock gene, PER1, were less sensitive to transcriptional inhibition than controls, and actually undercompensated for changes in temperature, resulting in shorter periods at higher temperatures.
These observations represent an advance in our understanding of how the clockwork integrates with cellular function, but should also facilitate enquiry into what additional mechanisms are responsible for maintaining rhythmicity, even when the control of gene expression has been impaired. It has been suggested earlier that post-translational mechanisms such as phosphorylation (e.g. casein kinase 1δ/ɛ and protein phosphatase 1 regulating PER2 degradation), and other cytosolic signalling processes, might have an important function in this context (Hastings et al, 2008). This paper will, therefore, further encourage a reappraisal of the contributions that various molecular mechanisms make to sustain cellular rhythms.
The authors also suggest that the processes underlying circadian temperature compensation may be shared with those that buffer the cell against the changes in gross transcriptional activity observed between different tissues. The molecular characterisation of such a mechanism would likely constitute a major challenge and a major advance for cell biology research. Furthermore, the possibility that PER1 constitutes a nexus between these two compensatory mechanisms serves to highlight the multi-functional roles of clock genes, especially when considering that the PER genes have also been implicated in tumour suppression (Lee, 2006).
Given that so many biological systems are under circadian regulation, this work should have profound implications for researchers in diverse fields. It is further hoped that these findings will be extended to other tissues, such as the hypothalamic suprachiasmatic nuclei—the conserved master clock in mammals—so that we can better understand how such compensatory mechanisms contribute to physiological and metabolic homeostasis.
References
- Akman OE, Locke JC, Tang S, Carre I, Millar AJ, Rand DA (2008) Isoform switching facilitates period control in the Neurospora crassa circadian clock. Mol Syst Biol 4:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Sage D, Unser M, Bauer C, d'Esmond T, Naef F, Schibler U (2009) Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J 28: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Hida A, Anderson DA, Izumo M, Johnson CH (2007) Cycling of cryptochrome proteins is not necessary for circadian-clock function in mammalian fibroblasts. Curr Biol 17: 1091–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger DB, Peskin CS (2003) A detailed predictive model of the mammalian circadian clock. Proc Natl Acad Sci USA 100: 14806–14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U (2004) The mammalian circadian timing system: from gene expression to physiology. Chromosoma 113: 103–112 [DOI] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, O'Neill JS (2008) Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol 18: R805–R815 [DOI] [PubMed] [Google Scholar]
- Lee CC (2006) Tumor suppression by the mammalian period genes. Cancer Causes Control 17: 525–530 [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ III, Takahashi JS, Kay SA (2007) Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH (2008) cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320: 949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, Kyriacou CP, Hastings MH (2006) Circadian orchestration of the hepatic proteome. Curr Biol 16: 1107–1115 [DOI] [PubMed] [Google Scholar]