EMBO J 28, 99–111 (2009); published online 21 January 2009
Covalent post-translational modifications of histones have important functions in transcription, replication, repair, and other aspects of eukaryotic chromosome dynamics. Trimethylation of lysine-4 on histone H3 is enriched at actively transcribed loci in many organisms. The impact of this histone modification on transcription has been extensively studied, but less is known about its effects on other chromosomal processes. An intriguing new study in this issue of EMBO Journal demonstrates that H3 lysine-4 trimethylation is critical in budding yeast for formation of the programmed DNA double-strand breaks that initiate homologous recombination during meiosis. These findings have important implications for elucidating the previously recognized but little understood connections between meiotic break formation and transcriptional promoters in this organism.
Eukaryotic genomes are packaged in a relatively compact form through incorporation of DNA into arrays of repeating nucleosomes. Each nucleosome consists of an octamer of histone proteins, usually two H2A–H2B dimers and an (H3-H4)2 tetramer, wrapped nearly twice around by ∼150 bp of DNA. One of the ways that cells control both the higher order folding of nucleosome arrays and the ability of other proteins to access the DNA in chromatin is through the placement and removal of covalent modifications on histones. One such modification is trimethylation of the lysine-4 residue of histone H3 (H3K4me3). This modification is enriched around the promoters and 5′ ends of actively transcribed genes. In the budding yeast Saccharomyces cerevisiae, H3K4 trimethylation is catalysed by the evolutionarily conserved Set1/COMPASS methyltransferase (also known as KMT2) associated with the RNA polymerase II transcriptional machinery (Shilatifard, 2008). H3K4me3 is thought to be involved in nucleosome remodelling and transcriptional elongation. In this issue of EMBO Journal, a new link is exposed between H3K4me3 and the formation of DNA double-strand breaks (DSBs) during meiosis (Borde et al, 2009).
Meiosis is the specialized cell division that reduces the genome complement by half to generate cells for sexual reproduction. During meiosis in most organisms, homologous maternal and paternal chromosomes pair and recombine with one another so that they can be accurately segregated during the first meiotic division (Hunter, 2006). Recombination also increases genetic diversity of progeny by breaking up chromosomal linkage groups. Meiotic recombination is initiated by programmed DSBs catalysed by the Spo11 transesterase. Interestingly, meiotic DSBs show a discontinuous distribution throughout the genome: regions that exhibit a heightened propensity to undergo breaks are referred to as DSB ‘hotspots'. In S. cerevisiae, such hotspots most often occur within promoter regions, consistent with the interpretation that DSB formation is facilitated by the open chromatin state characteristic of yeast promoters. Not all promoters are subject to significant DSB formation, however, and current data do not support a direct relationship between the level of transcription and formation of DSBs (Hunter, 2006).
In this issue, Borde et al (2009) demonstrate that deletion of the Set1 methyltransferase in S. cerevisiae leads to a dramatic decrease in the number of DSBs at the hottest hotspots examined, with 70% of these sites exhibiting a greater than two-fold reduction. This finding prompted the authors to more closely investigate the occurrence of H3K4me3 at these DSB sites. Using chromatin immunoprecipitation, they found that this modification was indeed highly enriched at DSB hotspots in promoters. Interestingly, the correlation between the presence of H3K4me3 and DSB hotspot activity was independent of the steady-state level of RNA from the genes. This result suggests that the connection between this histone modification and DSB formation is not tied directly to the transcriptional status. Moreover, H3K4me3 enrichment at hotspots is already seen during vegetative growth, that is, before cells enter meiosis, indicating that this modification is a pre-existing mark for regions that are especially permissive for DSB formation.
This study provides new insight into chromatin features that influence the distribution of meiotic recombination events. These results also tie in nicely with a previous analysis of H2B ubiquitination (H2Bub), which showed that preventing this modification lead to an overall decrease in DSBs (Yamashita et al, 2004). H2Bub promotes Set1/COMPASS activity and thus the appearance of H3K4me3 (Dover et al, 2002; Sun and Allis, 2002). Results of Borde et al (2009) may thus indicate that H2Bub promotes DSB formation indirectly through trimethylation of H3K4 (Figure 1). It will be interesting to determine whether other instances of known cross-talk among histone modifications also influence DSB formation. For example, presence of H3K4me3 is mutually exclusive with dimethylation of arginine-2 of histone H3 (H3R2me2), which is a modification primarily found in heterochromatin and inactive genes (Kirmizis et al, 2007). Investigation of whole-genome localization of H2Bub and H3R2me2 during meiotic prophase, and tests of whether H3R2me2 status influences DSB formation are thus of interest. Also, it should be noted that Borde et al (2009) limited their analysis of H3K4me3 distribution to only the ‘hottest' hotspots in the genome, that is, those which were five-fold or greater over background. Thus, it will also be interesting to examine to what extent the observed patterns also apply to the large number of much weaker DSB sites in the genome.
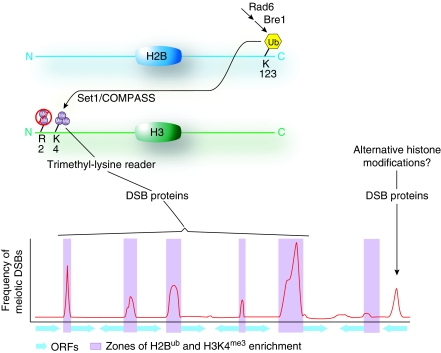
Figure 1.
Integration of multiple histone modifications promotes the formation of meiotic DSBs. It is likely that a combination of histone modifications is established at promoter regions prior to entry into meiosis. Ubiquitination of lysine-123 of H2B promotes subsequent Set1 activity upon lysine-4 of H3. H3K4me3 is then read by a putative trimethyl-lysine reader either contained as part of the pol II machinery or as a component of the DSB protein complex allowing for the formation of the majority of DSBs within promoters. Other subsets of DSBs occur within alternative chromatin environments and occur within ORFs, intergenic, and heterochromatic regions. Frequencies of meiotic DSBs are depicted by a smoothed histogram along a fictitious segment of yeast chromosome. Purple shading signifies promoters with nucleosomes enriched with the combination of H2Bub and H3K4me3, whereas ORFs are indicated by blue arrows.
The study of Borde et al (2009) joins a growing body of work revealing the influence of histone modifications on DSB formation in several organisms. Other recent examples include the demonstration of connections between DSB formation and proteins that control H3 lysine-9 methylation and H4 lysine-16 acetylation (which are marks associated with heterochromatin) and H3 lysine-36 methylation (which is found within the body of actively transcribed genes) (Reddy and Villeneuve, 2004; Mieczkowski et al, 2007; Merker et al, 2008). Interestingly, deletion of the histone acetyltransferase gene gcn5+ in Schizosaccharomyces pombe leads to a decrease in recombination at a synthetic DSB hotspot that is heavily acetylated in wild-type cells (Yamada et al, 2004). The synthesis of these findings supports a framework for meiotic DSB formation in which certain combinations of histone modifications create an environment favourable to DSB formation. It is clear, however, that different histone modifications influence particular hotspots to different extent. For example, although set1 deletion reduced or eliminated a majority of DSBs across the genome, 22 DSB sites were refractory to this decrease, even showing an increase in DSBs in the set1 mutant (Borde et al, 2009). These refractory sites lacked H3K4me3 in wild-type cells. These findings dramatically illustrate the fact that not all DSB sites are created equal, and that the ‘rules' that dictate hotspot activity differ from one hotspot to another (Petes, 2001).
How then is the DSB-forming machinery influenced by H3K4me3 and other modifications? One possibility is that one or more of the proteins required for DSB formation ‘reads' H3K4me3 by direct binding, thereby recruiting Spo11 and/or other factors to sites enriched for this modification. In S. cerevisiae, nine proteins in addition to Spo11 are required to generate DSBs, but the specific biochemical functions of these proteins are not well understood (Hunter, 2006). None of these proteins contains obvious motifs implicated in binding histone modifications (e.g. tudor domains or PHD fingers for binding methyl-lysine or bromodomains for binding acetyl-lysine), but available data do not exclude the possibility of such binding activities. A second possibility is that Spo11 or an associated factor interacts with other proteins that are dedicated ‘readers' of histone marks. For example, several proteins recognize H3K4me3 and are involved in transcription (Shilatifard, 2008). One of these, Chd1, is part of the Gcn5-containing SAGA acetyltransferase complex, which itself interacts with RNA pol II (Baker and Grant, 2007). The presence of RNA pol II or pol II-associated complexes at promoters might recruit the DSB machinery directly through protein–protein interactions or indirectly through effects on the ‘openness' of the chromatin. Interestingly, a direct binding partner of Spo11 is the WD-repeat protein Ski8, which has recently been reported to interact physically with the pol II subunit Rpo26 (Tarassov et al, 2008).
Elucidating the factors that control where meiotic DSBs occur is important for understanding the mechanism of DSB formation itself as well as for understanding the impact of meiotic recombination on genome structure and evolution. The findings of Borde et al (2009) emphasize the importance of histone modifications in facilitating DSB formation and open doors into an exploration of the complex and combinatorial modifications that function in this process.
References
- Baker SP, Grant PA (2007) The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 26: 5329–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A (2009) Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J 28: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Hunter N (2006) Meiotic recombination. In Molecular Genetics of Recombination, Aguilera A, Rothstein R (eds). Heidelberg: Springer-Verlag [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T (2007) Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449: 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker JD, Dominska M, Greenwell PW, Rinella E, Bouck DC, Shibata Y, Strahl BD, Mieczkowski P, Petes TD (2008) The histone methylase Set2p and the histone deacetylase Rpd3p repress meiotic recombination at the HIS4 meiotic recombination hotspot in Saccharomyces cerevisiae. DNA Repair (Amst) 7: 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Dominska M, Buck MJ, Lieb JD, Petes TD (2007) Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104: 3955–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2: 360–369 [DOI] [PubMed] [Google Scholar]
- Reddy KC, Villeneuve AM (2004) C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452 [DOI] [PubMed] [Google Scholar]
- Shilatifard A (2008) Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20: 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z-W, Allis CD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW (2008) An in vivo map of the yeast protein interactome. Science 320: 1465–1470 [DOI] [PubMed] [Google Scholar]
- Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K (2004) Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J 23: 1792–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Shinohara M, Shinohara A (2004) Rad6–Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci USA 101: 11380–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]