Abstract
Background
Escherichia coli induces heat shock genes to the temperature up-shift, and changes the metabolism by complicated mechanism. The heat shock response is of practical importance for the variety of applications such as temperature-induced heterologous protein production, simultaneous saccharification and fermentation (SSF) etc. However, the effect of heat shock on the metabolic regulation is not well investigated. It is strongly desired to understand the metabolic changes and its mechanism upon heat shock in practice for the efficient metabolite production by temperature up-shift. In the present research, therefore, we investigated the effect of temperature up-shift from 37°C to 42°C on the metabolism in view of gene expressions.
Results
The results of aerobic batch and continuous cultivations of E. coli BW25113 indicate that more acetate was accumulated with lower biomass yield and less glucose consumption rate at 42°C as compared to the case at 37°C. The down- regulation of the glucose uptake rate corresponds to the down-regulation of ptsG gene expression caused by the up-regulation of mlc gene expression. In accordance with up-regulation of arcA, which may be caused by the lower oxygen solubility at 42°C, the expressions of the TCA cycle-related genes and the respiratory chain gene cyoA were down-regulated. The decreased activity of TCA cycle caused more acetate formation at higher temperature, which is not preferred in heterologous protein production etc. This can be overcome by the arcA gene knockout to some extent. The time courses of gene expressions revealed that the heat shock genes such as groEL, dnaK, htpG and ibpB as well as mlc were expressed in much the same way as that of rpoH during the first 10–20 minutes after temperature up-shift. Under microaerobic condition, the fermentation changed in such a way that formate and lactate were more produced due to up-regulation of pflA and ldhA genes while ethanol was less produced due to down-regulation of adhE gene at higher temperature as compared to the case at 37°C.
Conclusion
The present result clarified the mechanism of metabolic changes upon heat shock from 37°C to 42°C based on gene expressions of heat shock genes, global regulators, and the metabolic pathway genes. It is recommended to use arcA gene knockout mutant to prevent higher acetate production upon heat shock, where it must be noted that the cell yield may be decreased due to TCA cycle activation by arcA gene knockout.
Background
Biological systems are known to be robust and adaptable to the culture environment. It became apparent that such robustness is inherent in the biochemical and genetic networks. Several genes that are necessary to respond to various environmental or nutritional changes require specific recognition by RNA polymerase associated with the alternative sigma factors such as σ32 [1], σE [2], σ54 [3], and σS [4]. In particular, the organisms respond to a sudden temperature up-shift by increasing the synthesis of a set of proteins. This phenomenon is called the heat shock response. The research on heat shock response of a microorganism contributes to the variety of practical applications such as temperature-induced heterologous protein production [5,6], simultaneous saccharification and fermentation (SSF) [7] etc.
The heat shock proteins play roles in the assembly and disassembly of macromolecular complex such as GroE [8], the intracellular transport such as Hsp70 [9], transcription such as σ70 [10], proteolysis such as Lon [11], and translation such as lysil tRNA synthetase [12]. The heat shock response in E. coli is mediated by Eσ32 [13], and it has been known that at least 26 genes are induced by heat shock [14], where E denotes the RNA polymerase holoenzyme. Among them, groEL, dnaK, and htpG are the genes which code for the major chaperones such as Hsp 60, Hsp 70, and Hsp 90. The ClpP, Lon and HtrC are involved in proteolysis. DnaK, DnaJ, and GrpE and RpoH are involved in the autoregulation of heat shock response [15-18]. It has been known that DnaK prevents the formation of inclusion bodies by reducing aggregation and promotion of proteolysis of misfolded proteins [19]. A bi-chaperone system involving DnaK and ClpB mediates the solubilization or disaggregation of proteins [20]. GroEL operates protein transit between soluble and unsoluble protein fractions and participates positively in disaggregation and inclusion body formation. Small heat shock proteins such as IbpA and IbpB protect heat- denatured proteins from irreversible aggregation and have been found to be associated with inclusion bodies [21,22].
Although many papers have been reported on the molecular mechanisms of heat shock proteins [23,24], very few researches have been reported on the effect of heat shock on the metabolism. Hoffmann et al. [6] investigated the metabolic adaptation of E. coli during temperature-induced recombinant protein production, and showed that cAMP/Crp-controlled LpdA of the pyruvate dehydrogenase complex(PDHc) and SdhA in the TCA cycle were induced four times, reaching a maximum at 1 h after temperature up-shift. It was also shown that the TCA cycle enzymes such as IcdA and Mdh were initially less produced but regained to their respective preshift values about 30 min after the temperature up-shift. More recently, Gadgil et al. [25] investigated the effect of temperature down-shift from 37°C to 33 and 28°C on gene expressions in E. coli. This kind of investigation is useful in analyzing the metabolic changes and investigating the effects of gene modification for strain improvement [26].
In the present study, we investigated how gene expression pattern changes in E. coli for the temperature up-shift from 37 to 42°C in relation to fermentation characteristics. The genes considered in the present study are listed in Additional file 1. The global regulators and their regulated genes are summarized in Additional file 2. Moreover, we investigated the dynamics of the expressions of such genes as heat shock genes, global regulators, and metabolic pathway genes to clarify the metabolic regulation upon heat shock.
Results
The batch cultivation results at two different temperatures of 37°C and 42°C indicate that the specific glucose consumption rate was lower at 42°C than at 37°C (data not shown). The maximum acetate concentration was 2.91 g/l at 42°C. Acetate was little consumed even after the glucose was depleted, whereas maximum acetate concentration was 2.69 g/l at 37°C. Acetate was consumed after the glucose was depleted at 37°C. The final cell concentration was lower at 42°C as compared to that at 37°C (3.32 and 3.92 g/l, respectively).
In order to make clear this phenomenon, aerobic continuous cultivation was conducted at the dilution rate of 0.2 h-1, where Table 1 shows the effect of culture temperature on the fermentation parameters. Table 1 indicates that acetate was more accumulated (p < 0.05), the cell yield was lower (p < 0.05), and the specific glucose consumption rate was lower (p < 0.1) at 42°C as compared to the case at 37°C, which is consistent with the above mentioned batch data.
Table 1.
Fermentation parameters for the aerobic chemostat culture of the wild type E. coli BW25113 at the dilution rate of 0.2 h-1
| Fermentation parameters | Culture temperature | % changes | |
| 37°C | 42°C | ||
| Specific glucose uptake rate (mmol/gDCW/h) | 2.46 ± 0.05 | 2.28 ± 0.04 | - 7.31 |
| Specific acetate production rate (mmol/gDCW/h) | 0.18 ± 0.02 | 0.32 ± 0.04 | +77.8 |
| Specific CER (mmol/gDCW/h) | 5.83 ± 0.05 | 5.87 ± 0.04 | + 0.69 |
| Specific OUR (mmol/gDCW/h) | 5.74 ± 0.06 | 5.76 ± .04 | + 0.35 |
| Biomass yield (gDCW/g substrate | 0.42 ± 0.01 | 0.34 ± 0.02 | - 19.0 |
± Standard deviation of three independent experiments.
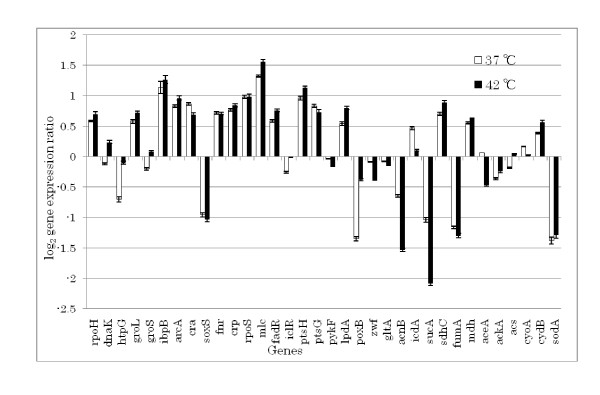
Figure 1a compares the gene expressions for the two different culture temperatures in the continuous culture (Table 1), where Fig. 1b depicts the changes in gene expressions on the metabolic pathways. Note that the samples were taken after the steady state was confirmed after the temperature was raised to 42°C. Figure 1 a indicates that the expression of rpoH was up-regulated (p < 0.10), and the expressions of dnaK, groL, groS, htpG, and ibpB were up-regulated (p < 0.1, p < 0.1, p < 0.05, p < 0.05 and p < 0.05, respectively), which are known to be under control of sigma factor (σ32). Figure 1 also shows the up-regulation of arcA gene expression (p < 0.1), where arcA gene product functions as a repressor of such genes as involved in the TCA cycle under microaerobic condition (Additional file 2) [27]. Figure 1 indicates that some of the TCA cycle and glyoxylate pathway genes such as icdA, sucA and aceA were down-regulated (p < 0.1, p < 0.1 and p < 0.05, respectively). Those genes are known to be under control of ArcA (arcA gene product) (Additional file 2). Figure 1 also indicates that the expressions of the respiratory chain genes such as cydB was up-regulated (p < 0.1), whereas cyoA was down regulated (p < 0.1). This may be due to the up-regulation of arcA and its gene product ArcA, since cydB operon is under the positive control, while cyoA is under negative control of ArcA (Additional file 2). Figure 1 also indicates that the expression of crp gene, which codes for cAMP receptor protein Crp, was up-regulated (p < 0.1), and the expression of lpdA, which is known to be under control of Crp (Additional file 2), was also up-regulated (p < 0.1) [6]. Moreover, mlc (making larger colonies) gene expression was higher, and the ptsG gene expression was lower (p < 0.05). Figure 1 also shows that cra (catabolite repressor activator) gene expression was down-regulated (p < 0.1), where cra gene product regulates the carbon flow in such a way that the gluconeogenesis was activated, whereas the glycolysis was repressed (Additional file 2). The gene expressions of fadR and iclR were also higher, where FadR activates iclR, and IclR is known to repress aceBAK.
Figure 1.
Effect of temperature up-shift on gene expressions in E. coli BW25113 under aerobic continuous culture at the dilution rate of 0.2 h-1 (a) comparison of gene expressions (The gene expressions are given as relative values to that of dnaA) (b) change in gene expressions on the metabolic pathways (red represents up-regulation, blue represents down-regulation, and black represents no change).
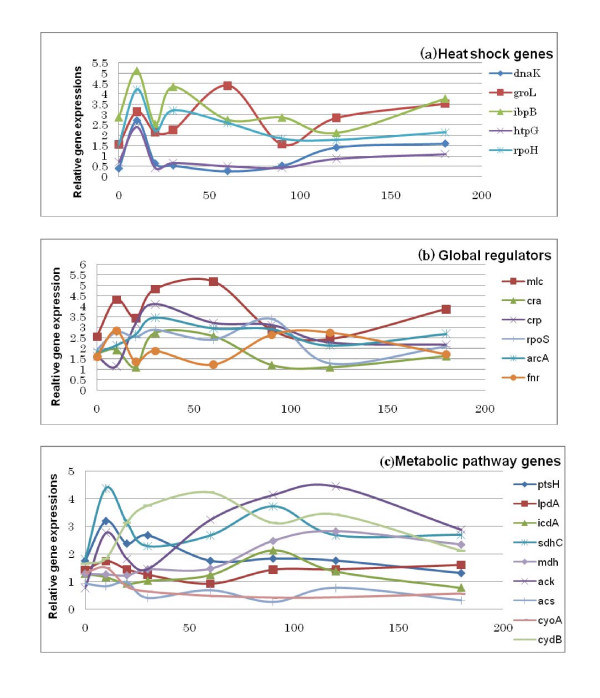
It has been known that the gene network forms a circuit so that the metabolic system is robust against external stimuli such as heat shock. Figure 2a shows how rpoH and the heat shock protein genes responded to the temperature up-shift with respect to time. The expression of rpoH gene shows an overshoot phenomenon during the initial 10–20 min, where this may be due to the feedback in the gene network. Moreover, the expressions of dnaK, groL, ibpB, htpG followed the same pattern as that of rpoH, which implies that those genes are under control of rpoH or σ32.
Figure 2.
Responses of gene expressions of E. coli BW25113 for the temperature up-shift (37°C to 42°C) under aerobic continuous culture at the dilution rate of 0.2 h-1. heat shock genes (b) global regulators (c) metabolic pathway genes.
Figure 2b shows the dynamic behavior of the global regulators, where it indicates that the expressions of mlc, crp, arcA tended to increase after the temperature up-shift, where it has been reported that the promoter regions of mlc and crp genes have the binding site for rpoH [28,29], whereas there is no such report on arcA.
Figure 2c shows the dynamic behavior of the metabolic pathway gene expressions, where ptsH and sdhC expressions became high at 10 min after the temperature up-shift but eventually settled down to lower values. Although sdhC gene expression shows little change in Fig. 1, Fig. 2c shows an increase in its expression level during the first 10 minutes after the temperature up-shift [6]. On the other hand, other TCA cycle genes such as mdh and icdA showed decrease expression during the first 30 mins [6]. The ackA gene expression tended to increase while acs expression tended to decrease, which may have partly caused the acetate accumulation. The expression of the respiratory chain gene such as cydB increased, while cyoA gene expression decreased.
Figures 1 and 2 indicate that arcA gene expression increased (p < 0.1), and the expressions of such genes as those under control of ArcA (Additional file 2) changed accordingly. Since arcA is not known to be under control of rpoH, the change in arcA gene expression may be indirect due to the lower dissolved oxygen concentration caused by the temperature up-shift. In order to see this in more detail, we cultivated arcA gene knockout mutant. Table 2 shows the fermentation parameters for the continuous cultivations of arcA mutant at two different temperatures. If we compare the results of Tables 1 and 2, the specific glucose consumption rate was higher, the specific acetate production rate was lower, and the cell yield was lower for the case of arcA mutant as compared to each case of 37°C and 42°C of the wild type strain. Table 2 also indicates that the acetate production rate was higher at 42°C as compared to the case at 37°C, where similar trend can be seen in Table 1.
Table 2.
Fermentation parameters for the aerobic chemostat culture of arcA mutant at the dilution rate of 0.2 h-1
| Fermentation parameters | Culture temperature | % changes | |
| 37°C | 42°C | ||
| Specific glucose uptake rate (mmol/gDCW/h) | 2.62 ± 0.06 | 2.48 ± 0.04 | - 5.34 |
| Specific acetate production rate (mmol/gDCW/h) | 0.11 ± 0.05 | 0.16 ± 0.07 | + 45.5 |
| Specific CER (mmol/gDCW/h) | 5.98 ± 0.05 | 6.22 ± 0.04 | + 4.01 |
| Specific OUR (mmol/gDCW/h) | 5.87 ± 0.06 | 6.11 ± 0.05 | + 4.08 |
| Biomass yield (gDCW/g substrate | 0.40 ± 0.01 | 0.35 ± 0.02 | - 12.5 |
± Standard deviation of three independent experiments.
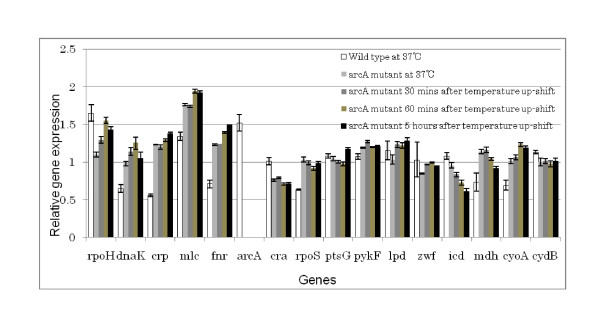
Figure 3 shows how gene expressions of arcA mutant changed at 30 min, 60 min, and 5 hours after temperature up-shift as compared to the case at 37°C as well as those of the wild type strain (white bar). By comparing the gene expressions of the wild type and arcA mutant at 37°C, it can be seen that mdh and cyoA genes were up-regulated, while cydB gene was down-regulated for arcA mutant. Figure 3 also indicates that dnaK gene expression followed the same time profile as that of rpoH. The crp and mlc gene expressions became higher as time proceeded. Figure 3 also indicates that fnr gene expression increased, whereas cra gene expression decreased, and icdA and mdh gene expressions eventually decreased.
Figure 3.
Effect of temperature on gene expressions of arcA gene knockout mutant under aerobic continuous culture at the dilution rate of 0.2 h-1.
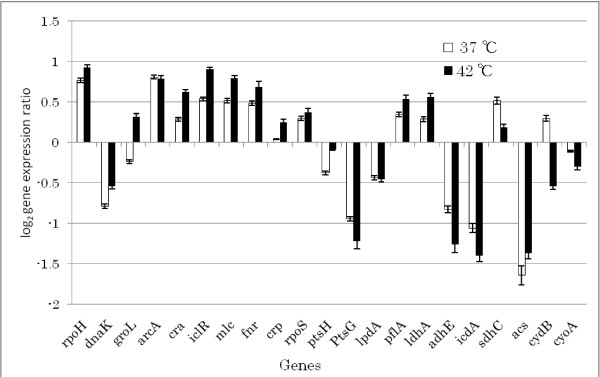
In order to see the effect of aeration, we have also conducted batch cultivations at microaerobic (or essentially anaerobic) condition. Table 3 shows the comparison of the yields at the time when glucose was depleted in the batch culture. Table 3 indicates that acetate, formate and lactate were more produced, while ethanol was less produced at 42°C as compared to the case at 37°C. Figure 4 indicates that rpoH gene expression increased at higher temperature, and the expressions of such genes as dnaK and groL followed the same pattern. The arcA gene expression changed little but fnr gene expression increased at higher temperature. The gene expressions of cra, iclR and crp increased at higher temperature. The expressions of ptsG, icdA, sdhC, adhE and cyo genes decreased, while pflA and ldhA expressions increased at higher temperature as compared to those at 37°C, which is consistent with the fermentation data of Table 3.
Table 3.
Fermentation parameters for the microaerobic batch culture of wild type Escherichia coli.
| Growth parameters | Culture temperature | % changes | |
| 37°C | 42°C | ||
| Biomass yield (gDCW/g substrate) | 0.12 ± 0.01 | 0.098 ± 0.02 | - 18.3 |
| Acetate yield (g/g substrate) | 0.25 ± 0.03 | 0.32 ± 0.02 | + 28.0 |
| Lactate yield (g/g substrate) | 0.26 ± 0.02 | 0.31 ± 0.01 | + 19.2 |
| Formate yield (g/g substrate) | 0.25 ± 0.03 | 0.29 ± 0.02 | + 16.0 |
| Succinate yield (g/g substrate) | 0.02 ± 0.002 | 0.03 ± .003 | + 50.0 |
| Ethanol yield (g/g substrate) | 0.15 ± 0.01 | 0.13 ± 0.02 | -1 3.3 |
± Standard deviation of three independent experiments.
Figure 4.
Effect of temperature on gene expressions of E. coli BW 25113 under microaerobic condition.
Discussion
To survive, cells have to control gene expressions precisely in response to the changes in the growth environment. The microorganism such as E. coli attains this primarily at the transcription level. To control the initiation of the specific transcription, the cell uses diverse mechanisms including various sigma factors. The classical heat shock regulon has been shown to be under the control of σ32 transcription factor, the product of the rpoH gene [30]. The regulation of the sigma factor (σ32) is complex and depends on the feedback control loops involving the dnaK chaperone and temperature-induced changes in mRNA secondary structure [31]. The relative levels of the major heat shock genes such as dnaK, groS, groL, ibpB, lonA and htpG were found to be up-regulated after the temperature up-shift. The expressions of heat shock genes such as dnaK, groL, and ibpB increased in the early induction phase (first 10–20 minutes) and then declined (Fig. 2a). In E. coli, heat shock protein synthesis rates peak at about 5~10 min after the temperature up-shift and then declined to a new steady-state levels [32]. The heat shock response is made transcriptionally, where it has been known that the RNA polymerase core (E) binds to new initiation subunit σ32 [33], and the resulting holoenzyme Eσ32, transcribes only heat shock genes [1], which have promoter sequences that differ from those transcribed by E plus σ70, the normal vegetative initiation factor [34]. The transcription factor σ70 is itself a heat shock protein and the increase in its concentration after heat shock may contribute to its decline in heat shock protein synthesis. Moreover, other heat shock proteins, in particular the dnaK gene product contributes to the shutoff, since the mutations in their genes prolong the high level synthesis of heat shock proteins [35]. The heat shock response must be tightly regulated in order to allow rapid changes in heat shock protein synthesis rates. Although the level of mRNA transcribed from the rpoH gene increases after heat shock, their increases may be insufficient and too slow to be the sole explanation of the rapid effect of the heat shock. It has been shown that the concentration of active σ32 limits the expression of heat shock genes, and that the stability of σ32 is modulated [32].
Because of the rapid turn-over (half life of less than 1 min), the cellular concentration of σ32 is very low at normal temperature and is limiting for the transcription of the heat shock gene. Upon temperature up-shift, σ32 becomes transiently stabilized until the beginning of the shut-off phase of the heat shock response. The heat shock response is induced as a consequence of declining σ32 levels and inhibition of σ32 activity. Stress- dependent changes in heat shock gene are mediated by the antagonistic action of σ32 and negative modulators which act upon σ32. These modulators are the DnaK chaperone system which inactivates σ32 by direct association and mediates its degradation by proteases [36]. Degradation of σ32 is mediated mainly by FtsH, and ATP dependent metallo- protease within the inner membrane. The heat shock proteins increased immediately after the temperature up-shift, reached a maximum 5–15 min later, and decreased to preshift values largely within 1 h, while the maximum induction of many heat-shock proteins including DnaK and HtpG reached at least 30 min later.
The E. coli cyclic AMP (cAMP) receptor protein Crp activates transcription for more than 100 promoters. When bound to its allosteric effector cAMP, the Crp homodimer binds to the specific DNA sites near target promoters, enhancing the binding of RNA polymerase holoenzyme (RNAP), and facilitating the initiation of the transcription. The present result indicates that crp gene expression increased and lpdA gene expression followed the similar pattern, but the effect of these gene expressions may not be significant for the change in the metabolism, or not clear at this time.
The present result shows that mlc gene expression followed the same pattern as that of rpoH upon heat shock, which confirms that Eσ32 is involved in the expression of mlc gene. It has been shown that Eσ32 plays an important role in balancing the relative concentration of Mlc and EIICB in response to the availability of glucose in order to maintain inducibility of Mlc regulon at higher temperature [28]. When Mlc was overproduced, it has been known to reduce acetate accumulation [37], and causes slow growth but gives better performance for recombinant protein production [38]. Mlc is a global regulator of carbohydrate metabolism, and regulates the expression of pts operon (Additional file 2). It has been known that Mlc represses manXYZ encoding enzyme II of the mannose PTS [39], malT encoding the activator of maltose operon, and mlc itself negatively [40]. Moreover, ptsG encoding enzyme IICB of the glucose PTS (EIICBglu) and the pts operon encoding general PTS proteins are also known to be repressed by Mlc [41,42]. The mlc promoter is very weak because nucleotide sequence of -10 region of the promoter differs from the consensus sequence of the strong promoter of E. coli. In addition, Mlc expression is autoregulated by Mlc itself. Therefore, the intracellular concentration of Mlc is limited in E. coli [43]. The mlc gene has been known to be transcribed by two promoters, P1 and P2, and have a binding site of its own gene product. It has been shown by in vitro transcription assays of mlc gene that P2 promoter could be recognized by RNA polymerase containing the heat shock sigma factor σ32 (Eσ32) as well as Eσ70, while P1 promoter is only recognized by Eσ70. Figures 1 and 4 indicate that ptsG gene expression decreased (p < 0.05) in accordance with the up-regulation of mlc gene expression, while ptsH did not follow such pattern, where ptsH is not under control of Mlc and it was activated partly due to down-regulation of cra gene expression (Additional file 2).
Let us consider the production mechanism of acetate at higher temperature. In the typical batch cultivation, the cells must switch efficiently from the rapid growth on a favored carbon source such as glucose to a much slower growth on the excreted by-products such as acetate. Acetate excretion occurs through the phosphotransacetylase-acetate kinase (Pta-Ack) pathway, or may possibly by Pox (pyruvate oxidase) pathway. Acetate utilization occurs through AcCoA synthetase (Acs). This high- affinity acetate- scavenging enzyme converts acetate to AcCoA, where cells introduce it into the TCA cycle to generate energy and/or the glyoxylate pathway to build cell constituents. The higher expression of acs accelerates acetate assimilation in the presence of acetate [44,45], which leads to the activation of glyoxylate pathway. Transcription occurs from two σ70- dependent promoters such as the distal promoter acs P1 and proximal promoter acs P2 [44,46]. While multiple factors influence transcription, Crp appears to function directly as the critical transcription factor. Cells control this acetate switch primarily by controlling the initiation of acs transcription from the major promoter acsP2 [44,47]. Activation of acs transcription depends on the cAMP-Crp. The cAMP-Crp binds two sites within the acs regulatory region. However, it has been shown that Fis and Ihf independently modulate Crp- dependent activation of acsP2 transcription [48], and the mechanism is not so simple. As such, the activation of crp may cause acs to be up-regulated (Figs. 1 and 4). The acs gene is also under control of rpoS. In our previous investigation, it was shown that acs was expressed in an rpoS- dependent manner during different phases of the batch growth [49], but rpoS did not change much in the present case as shown in Figs. 1 and 4.
Although cellular ATP may increase for short period after the temperature up-shift in E. coli [50], it eventually decreases at higher temperature [6,50]. It has also been reported that the specific CO2 production rate as well as O2 consumption rate increased upon temperature up-shift [6,50,51]. As a result, the cell yield decreased and the cell maintenance increased [6,52]. Although it has been reported that the TCA cycle flux increased upon temperature up-shift at the specific growth rate of 0.08 h-1[50], recent investigation based on 13C- labeled experiment indicates that the TCA cycle flux became low at the dilution rates of 0.45 and 0.32 h-1[53]. Our result implies the repression of TCA cycle genes due to up-regulation of arcA gene, while the respiratory activity became higher due to up-regulation of cyd gene, which may be caused by the up-regulation of arcA gene. The icdA and aceA genes are known to be repressed by ArcA/B and activated by Cra (Additional file 2). The up-regulation of arcA gene expression and down regulation of cra gene expression both acted to repress icdA and aceA genes, and thus the TCA cycle as well as the glyoxylate pathway was repressed. In accordance with this, the fadR and iclR expressions were also up-regulated, which are known to repress aceBAK operon. This may have caused more acetate accumulation. Moreover, the down-regulation of cyoA may have limited the respiratory activity, while it may be counteracted by the activation of cydB gene. It has been reported that the respiration was activated during the temperature up-shift [6]. It may be due to the activation of cydB gene expression since Km value is lower or the affinity to oxygen is higher for Cyd as compared to Cyo. The global regulator, arcA showed increased expression after the temperature up-shift (especially first 30 mins) and modulated the expressions of such genes as cydB, cyoA, icdA etc. The up-regulation of arcA gene may not be the direct effect of heat shock but indirectly due to lower dissolved oxygen concentration caused by the lower solubility at higher temperature [50].
The down-regulation of cra gene expression may not be direct but may be due to the increase in the glucose concentrations in the fermentor, since the glucose uptake rate was reduced at higher temperature (Table 1). It has been reported that superoxide dismutase gene (sod) is induced in response to the oxidative stress imposed by dioxygen or by the redox active compounds such as viologens or quinones caused by the temperature up-shift [54]. It has also been reported that the exposure of a sodA/B null mutant E. coli to aerobic heat stress caused a profound loss of viability [55]. However, our experimental data of Fig. 1 indicate little change in sod expression. This may be due to the fact that the temperature was changed from 37°C to 42°C in the present experiment, while above experiment[52] was conducted at 45–48°C. Moreover, the sod gene is under control of SoxRS, where it becomes significant under dual osmotic and heat stresses [56].
The common obstacle for the efficient metabolite production is the formation of by-products such as acetate. The present result suggests to use arcA gene knockout mutant. However, arcA gene knockout reduces the cell yield due to the activation of the TCA cycle. The cell yield can be increased by further knockout of such genes as fadR [57] or iclR [58]. In some cases, the dilution rate was lowered at the time of temperature up-shift in order to reduce the acetate formation, but the comparison must be careful in such a situation due to both effects of temperature up-shift and lower dilution rate. Moreover, in the case of induction of heterologous proteins, the result must be carefully interpreted, since both higher temperature and hetrologous protein production impose stress to the cell [59]. The present results do not contain such factors as different dilution rate or heterologous protein production, and it was clarified how the metabolism changed by the temperature up-shift in view of gene expressions. The overall mechanism may be illustrated as Fig. 5, where the left half part of heat shock regulation is already known [36], while the right half part was found in the present investigation.
Figure 5.
Schematic diagram of the effect of heat shock on gene expressions and metabolic regulation.
Conclusion
As shown in Fig. 5, the present research result clarified the mechanism of metabolic changes upon temperature up-shift from 37°C to 42°C based on gene expressions of heat shock genes, global regulators, and the metabolic pathway genes. In particular, the temperature up-shift caused the up-regulation of mlc gene, which in turn caused the down-regulation of ptsG gene expression, and finally caused the decreased in the specific glucose up-take rate. It was also found that the temperature up-shift also caused the up-regulation of crp gene expression, and activated lpdA gene expression. Moreover, it was shown that the temperature up-shift caused other TCA cycle genes to be repressed due to up-regulation of arcA gene indirectly caused by lower dissolved oxygen concentration and caused higher acetate production. This information is useful for the variety of applications such as temperature-induced heterologous protein productions, SSF etc. In particular, it is suggested to use arcA gene knockout mutant to reduce the acetate overproduction caused by the temperature up-shift with some careful consideration of its effect on the cell yield.
Materials and methods
Bacterial strains and culture conditions
The strains used were Escherichia coli BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78), and its arcA gene knockout mutant (JW4364). Batch and continuous cultivations were conducted using 2- l jar fermentor (M-100, Rikakikai Co. Tokyo, Japan), where the temperature was kept constant either at 37°C or 42°C, and the pH of the culture was maintained at 7.0 ± 0.1 by addition of 2.0 M HCl or 2.0 M NaOH with a pH controller. The air flow rate was maintained at 1 liter min-1 and the agitation speed was 350 rpm to ensure dissolved oxygen level to be about 40% of air saturation in the aerobic cultivation. The microaerobic cultivation was initiated by 2 h of aerobic cultivation followed by the microaerobic cultivation without supplying air and slowing down the agitation speed to around 100 rpm so that the cultivation was nearly anaerobic. Since the outlet of the fermentor was open to the atmosphere via the liquid bottle for mist trapping, the produced CO2 in the fermentor flowed out without increasing the overhead pressure. CO2 and O2 concentrations were measured by the off-gas analyzer (ABLE Co., Japan). The M9 synthetic medium was used, where it contained 10 g of glucose per liter, 48 mM Na2HPO4, 22 mM KH2PO4, 10 mM NaCl, and 30 mM (NH4)2SO4. The following components were filter sterilized and then added (per liter of final medium): 1 ml of 1 M MgSO4, 1 ml of 0.1 mM CaCl2, 1 ml of 1 mg of Vitamin B1 per liter, and 10 ml of trace element solution containing (per liter) 0.55 g of CaCl2, 1 g of FeCl3, 0.1 g of MnCl2.4H2O, 0.17 g of ZnCl2, 0.043 g of CuCl2.2H2O, 0.06 g of CoCl2.6H2O and 0.06 g Na2MoO4.2H2O. Continuous cultivations were performed at the dilution rate of 0.2 h-1, where the culture temperature was raised from 37°C to 42°C after the steady-state was ascertained.
Measurements of biomass and extracellular metabolite concentrations
Cell concentration was measured by the optical density (OD) of the culture broth at 600 nm wave length with a spectrophotometer (Ubet-30, Jasco Co., Tokyo, Japan), and then converted to dry cell weight (DCW) per liter based on the relationship between OD and DCW previously obtained. Glucose concentration was measured using enzymatic kit (Wako Co., Osaka, Japan). Acetate, formate, lactate, succinate, and ethanol concentrations were also measured using enzymatic kits (Boehringer Co., Mannheim, Germany).
RNA isolation, cDNA synthesis and PCR amplification
The 2.5 μl of culture broth was immediately suspended with 5 μl RNAprotect bacteria reagent. The samples were kept on ice for 5 minutes. After centrifugation at 10,000 rpm (4°C, 10 min), the supernatant was discarded and the pellet was stored at -80°C until RNA isolation. Total RNA was isolated from E. coli cells by Qiagen RNeasy Mini Kit (QIAGEN K.K., Japan) according to the manufacturer's recommendation. The quantity and purity of the RNA were determined by the optical density measurements at 260 and 280 nm and by 1% formaldehyde agarose gel electrophoresis. The sequences of the primers used in the present study are given elsewhere [60] except the followings:
mlc 5' AGCAGACCAACGCGGGCGCG 3'
5' GACTATACGCAGGAAGGGCC 3'
Criteria for the design of the gene-specific primer pairs were followed according to Sambrook and Russel [61]. The primers used in this study were synthesized at Hokkaido System Science Co. (Sapporo, Hokkaido, Japan). In all cases, the primer-supplied company confirmed the absolute specificity of the primers.
RT-PCR reactions were carried out in a TaKaRa PCR Thermal Cycler (TaKaRa TP240, Japan) using Qiagen One Step RT-PCR Kit (QIAGEN K.K., Japan). The reaction mixture was incubated for 30 min at 50°C for reverse transcription (cDNA synthesis) followed by 15 min incubation at 95°C for initial PCR activation. Then the process was subjected to 30 cycles of amplification which consisted of a denaturing step (94°C for 1 min), an annealing step (approximately 5°C below melting temperature, of primers for 1 min) and an extension step (72°C for 1 min), and finally the reaction mixture of 25 μl was subjected for 10 min at 72°C for final extension. To check for nucleic acid contamination, one negative control was run in every round of RT-PCR. This control lacks the template RNA in order to detect possible contamination of the reaction components. 5 μl of amplified products were run on 1.8% agarose gel. Gels were stained with 1 mg ml-1 of ethidium bromide, photographed using a Digital Image Stocker (DS-30, FAS III, Toyobo, Osaka, Japan) under UV light and analyzed using Gel-Pro Analyzer 3.1 (Toyobo, Osaka, Japan) software. In order to determine the optimal amount of input RNA, the two-fold diluted template RNA was amplified in RT-PCR assay under identical reaction condition to construct a standard curve for each gene product. When the optimal amount of input RNA was determined for each gene product, RT-PCR was carried out under identical reaction condition to detect differential transcript levels of genes. The gene dnaA, which encodes E. coli DNA polymerase and is not subjected to variable expression, i.e. abundant expression at relatively constant rate in most cells, was used as an internal control in the RT-PCR determinations. The gene expressions are given as relative values to that of dnaA. To calculate the standard deviation, RT-PCR was independently performed three times under identical reaction condition.
To ensure that the observed expression changes were statistically significant, the Student's t-test was applied.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Monirul Hasan: All experiments. Kazuyuki Shimizu: Experimental design and manuscript preparation.
Supplementary Material
Five tables.
Global regulators and its regulated genes.
Contributor Information
Chowdhurry Mohammad Monirul Hasan, Email: monirkyushu@gmail.com.
Kazuyuki Shimizu, Email: shimi@bio.kyutech.ac.jp.
References
- Grossman AD, Erickson JW, Gross CA. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- Erickson JW, Gross CA. Identification of the σE subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes & Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- Hirschman J, Wong PK, Sei K, Keener J, Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci USA. 1985;82:7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey MR, Loewen PC. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel sigma transcription factor. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockney RC. Recent developments in heterologous protein production in Escherichia coli. Trends Biotechnol. 1994;12:456–463. doi: 10.1016/0167-7799(94)90021-3. [DOI] [PubMed] [Google Scholar]
- Hoffmann F, Weber J, Rinas U. Metabolic Adaptation of Escherichia coli During Temperature-Induced Recombinant Protein Production: 1. Readjustment of Metabolic Enzyme Synthesis. Biotechnol Bioeng. 2002;80:313–319. doi: 10.1002/bit.10379. [DOI] [PubMed] [Google Scholar]
- Philippidis GP, Smith TK, Wyman CE. Study of the enzymatic hydrolysis of cellulose for production of fuel ethanol by the simultaneous saccharification and fermentation process. Biotechnol Bioeng. 2004;41:846–853. doi: 10.1002/bit.260410903. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Properties of a mutant of Escherichia coli in bacteriophage λ head formation (groE), II the propagation of phase λ. J Mol Biol. 1973;76:25–44. doi: 10.1016/0022-2836(73)90079-X. [DOI] [PubMed] [Google Scholar]
- Chirico WJ, Waters MG, Blobel G. 70 K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Taylor WE, Straus DB, Grossman AD, Burton ZF, Gross CA, Burgess RR. Transcription from a heat-inducible promoter causes heat shock regulation of the sigma subunit of E. coli RNA polymerase. Cell. 1984;38:371–381. doi: 10.1016/0092-8674(84)90492-6. [DOI] [PubMed] [Google Scholar]
- Goff SA, Casson LP, Goldberg AL. Heat shock regulatory gene htpR influences rates of protein degradation and expression of the lon gene in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:6647–6651. doi: 10.1073/pnas.81.21.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbogelen RA, Vaughn V, Neidhardt FC. Gene for heat-inducible lysyl tRNA synthetase (lys U) maps near cadA in Escherichia coli. J Bacteriol. 1983;153:1066–1068. doi: 10.1128/jb.153.2.1066-1068.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T, Nagai H, Mori H. Regulation of the heat shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- Chuang SE, Blattner FR. Characterization of Twenty Six New Heat Shock Genes of Escherichia coli. J Bacteriol. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Gross CA. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-Z. [DOI] [PubMed] [Google Scholar]
- Gamer J, Bujard H, Bukau B. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor σ32. Cell. 1992;69:833–842. doi: 10.1016/0092-8674(92)90294-M. [DOI] [PubMed] [Google Scholar]
- Liberek K, Galitski TP, Zylicz M, Georgopoulos C. The DnaK chaperone modulates the heat-shock response of Escherichia coli by binding to the σ32 transcription factor. Proc Natl Acad Sci USA. 1992;89:3516–3520. doi: 10.1073/pnas.89.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Spence J, Georgopoulos C. Modulation of stability of the Escherichia coli heat shock regulatory factor σ32. J Bacteriol. 1983;171:1585–1589. doi: 10.1128/jb.171.3.1585-1589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Friedrich KL, Schonfeld HJ, Vierling E, Bukau B. Refolding of substrates bound to small Hsps relies on a diaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem. 2003;278:31033–42. doi: 10.1074/jbc.M303587200. [DOI] [PubMed] [Google Scholar]
- Schlieker C, Bukau B, Mogk A. Prevention and reversion of protein aggregation by molecular chaperones in the E. coli cytosol: implications for their applicability in biotechnology. J Biotechnol. 2002;96:13–21. doi: 10.1016/S0168-1656(02)00033-0. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Miyakawa M, Matsumura Y, Tsuchido T. E. coli small heat shock proteins, IbpA and IbpB, protect enzymes from inactivation by heat and oxidants. Eur J Biochem. 2002;269:2907–17. doi: 10.1046/j.1432-1033.2002.02958.x. [DOI] [PubMed] [Google Scholar]
- Sorensen HP, Mortensen KK. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact. 2005;4:1. doi: 10.1186/1475-2859-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen R, Ron EZ. Proteome analysis in the study of the Bacterial Heat-shock response. Mass spect Rev. 2002;21:244–265. doi: 10.1002/mas.10031. [DOI] [PubMed] [Google Scholar]
- Richmond CS, Glasner DJ, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadgil M, Kapur V, Hu WS. Transcriptional Response of Escherichia coli to Temperature Shift. Biotechnol Prog. 2005;21:689–699. doi: 10.1021/bp049630l. [DOI] [PubMed] [Google Scholar]
- Kao CM. Functional genomics technologies: creating: new paradigms for fundamental and applied biology. Biotechnol Prog. 1999;15:304–311. doi: 10.1021/bp990027q. [DOI] [PubMed] [Google Scholar]
- Spiro S, Guest JR. Adaptive responses to oxygen limitation in Escherichia coli. Trends Biochem Sci. 1991;16:310–314. doi: 10.1016/0968-0004(91)90125-F. [DOI] [PubMed] [Google Scholar]
- Shin D, Lim S, Seok Y, Ryu S. Heat Shock RNA Polymerase (Eσ32) Is involved in the transcription of mlc and crucial for induction of the Mlc regulon by Glucose in Escherichia coli. J Biol Chem. 2001;276:25871–25875. doi: 10.1074/jbc.M101757200. [DOI] [PubMed] [Google Scholar]
- Ryu S. CRP.cAMP-dependent transcription activation of the Escherichia coli pts Po promoter by the heat shock RNA polymerase (Eσ32) in vitro. Mol Cells. 1998;8:614–617. [PubMed] [Google Scholar]
- Gross CA, Straus DB, Erickson JW, Yura T. The function and regulation of heat shock proteins in Escherichia coli. In: Morimoto RJ, et al, editor. Stress proteins in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y; 1990. pp. 167–189. [Google Scholar]
- Yura T, Nakahigashi K. Regulation of heat shock response. Curr Opin Microbial. 1999;2:153–158. doi: 10.1016/S1369-5274(99)80027-7. [DOI] [PubMed] [Google Scholar]
- Tilly K, Erickson J, Sharma S, Georgopoulos C. Heat shock regulatory gene rpoH mRNA level increases after heat shock in Escherichia coli. J Bacteriol. 1986;168:1155–1158. doi: 10.1128/jb.168.3.1155-1158.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly S, Colemant T, Fu CF, Brot N, Weissbach H. Correlation between the 32-kDa a factor levels and in vitro expression of Escherichia coli heat shock genes. Proc Nati Acad Sci. 1987;84:8365–8369. doi: 10.1073/pnas.84.23.8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowing DW, Bardwell JCA, Craig EA, Woolford C, Hendrix RW, Gross CA. Consensus Sequence for Escherchia coli Heat Shock Gene Promoter. Proc Natl Acad Sci USA. 1985;82:2679–2683. doi: 10.1073/pnas.82.9.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, McKittrick N, Zylicz M, Georgopoulos C. The dnaK protein modulates the heat-shock response of Escherichia coli. Cell. 1983;34:641–646. doi: 10.1016/0092-8674(83)90396-3. [DOI] [PubMed] [Google Scholar]
- Arsene F, Tomoyasu T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55:3–9. doi: 10.1016/S0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- Hosono K, Kakuda H, Ichihara S. Decreasing accumulation of acetate in a rich medium by Escherichia coli on introduction of genes on a multicopy plasmid. Biosci Biotechnol Biochem. 1995;59:256–261. doi: 10.1271/bbb.59.256. [DOI] [PubMed] [Google Scholar]
- Cho S, Shin D, Ji GE, Heu S, Ryu S. High-level recombinant protein production by overexpression of Mlc in Escherichia coli. J Biotechnol. 2005;119:197–203. doi: 10.1016/j.jbiotec.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol Microbiol. 1998;27:369–380. doi: 10.1046/j.1365-2958.1998.00685.x. [DOI] [PubMed] [Google Scholar]
- Kim SY, Nam TW, Shin D, Koo BM, Seok YJ, Ryu S. Purification of Mlc and Analysis of Its Effects on the pts Expression in Escherichia coli. J Biol Chem. 1999;274:25398–25402. doi: 10.1074/jbc.274.36.25398. [DOI] [PubMed] [Google Scholar]
- Kimata K, Inada T, Tagami H, Aiba H. A global repressor (Mlc) is involved in glucose induction of the ptsG gene encoding major glucose transporter in Escherichia coli. Mol Microbiol. 1998;29:1509–1519. doi: 10.1046/j.1365-2958.1998.01035.x. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kimata K, Inada T, Tagami H, Aiba H. Negative regulation of the pts operon by Mlc: Mechanism underlying glucose induction in Escherichia coli. Genes Cells. 1999;4:391–399. doi: 10.1046/j.1365-2443.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- Nam T, Cho S, Shin D, Kim J, Jeong J, Lee J, Roe J, Seok Y. The Escherichia coli glucose transporter enzyme IICBGlc recruits the global repressor Mlc. EMBO J. 2001;120:491–498. doi: 10.1093/emboj/20.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Beatty CM, Browning DF, Busby SJW, Simel EJ, Hovel-Miner G, Wolfe AJ. Regulation of Acetyl Coenzyme A Synthetase in Escherichia coli. J Bacteriol. 2000;182:4173–4179. doi: 10.1128/JB.182.15.4173-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Bennettb GN, San K-Y. Chemostat culture characterization of Escherichia coli mutant strains metabolically engineered for aerobic succinate production: A study of the modified metabolic network based on metabolite profile, enzyme activity, and gene expression profile. Metabolic Eng. 2005;7:337–352. doi: 10.1016/j.ymben.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Browning DF, Beatty CM, Wolfe AJ, Cole JA, Busby SJW. Independent regulation of the divergent Escherichia coli nrfA and acs P1 promoters by a nucleoprotein assembly at a shared regulatory region. Mol Microbiol. 2002;43:687–701. doi: 10.1046/j.1365-2958.2002.02776.x. [DOI] [PubMed] [Google Scholar]
- Beatty CM, Browning DF, Busby SJW, Wolfe AJ. Cyclic AMP Receptor Protein-Dependent Activation of the Escherichia coli acs P2 Promoter by a Synergistic Class III Mechanism. J Bacteriol. 2003;185:5148–5157. doi: 10.1128/JB.185.17.5148-5157.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Beatty CM, Sanstad EA, Gunn KE, Busby SJ, Wolfe AJ. Modulation of CRP-dependent transcription at the Escherichia coli acs P2 promoter by nucleoprotein complexes: anti-activation by the nucleoid proteins FIS and IHF. Mol Microbiol. 2004;51:241–254. doi: 10.1046/j.1365-2958.2003.03824.x. [DOI] [PubMed] [Google Scholar]
- Rahman M, Shimizu K. Altered acetate metabolism and biomass production in several Escherichia coli mutants lacking rpoS-dependent metabolic pathway genes. Mol Biosyst. 2008;4:160–9. doi: 10.1039/b712023k. [DOI] [PubMed] [Google Scholar]
- Soini J, Falschlehner C, Mayer C, Böhm D, Weinel S, Panula J, Vasala A, Neubauer P. Transient increase of ATP as a response to temperature up-shift in Escherichia coli. Microb Cell Fact. 2005;4:9. doi: 10.1186/1475-2859-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann F, Rinas U. Plasmid amplification in Escherichia coli after temperature up-shift is impaired by induction of recombinant protein synthesis. Biotechnol Lett. 2001;23:1819–1825. doi: 10.1023/A:1012718200638. [DOI] [Google Scholar]
- Weber J, Hoffmann F, Rinas U. Metabolic adaptation of Escherichia coli during temperature-induced recombinant protein production: 2. Redirection of metabolic fluxes. Biotechnol Bioeng. 2002;80:320–330. doi: 10.1002/bit.10380. [DOI] [PubMed] [Google Scholar]
- Wittmann C, Weber J, Betiku E, Krömera J, Böhmb D, Rinas U. Response of fluxome and metabolome to temperature-induced recombinant protein synthesis in Escherichia coli. J Biotechnol. 2007;132:375–384. doi: 10.1016/j.jbiotec.2007.07.495. [DOI] [PubMed] [Google Scholar]
- Privalle CT, Fridovich I. Induction of superoxide dismutase in Escherichia coli by heat shock. Proc Natl Acad Sci USA. 1987;84:2723–2726. doi: 10.1073/pnas.84.9.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benov L, Fridovich I. Superoxide Dismutase Protects against Aerobic Heat Shock in Escherichia coli. J Bacteriol. 1995;177:3344–3346. doi: 10.1128/jb.177.11.3344-3346.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera TS, Csonka LN, Paliy O. Genome wide Transcriptional Responses of Escherichia coli K-12 to Continuous Osmotic and Heat Stresses. J Bacteriol. 2008;190:3712–3720. doi: 10.1128/JB.01990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Shimizu K. Effect of fadR gene knockout on the metabolism of Escherichia coli based on analyses of protein expressions, enzyme activities and intracellular metabolite concentrations. Enzyme Microbial Technol. 2006;38:512–520. doi: 10.1016/j.enzmictec.2005.07.019. [DOI] [Google Scholar]
- Sarkar D, Siddiquee KAZ, Arauzo-Bravo MJ, Oba T, Shimizu K. Effect of cra gene knockout together with edd and iclR genes knockout on the metabolism in Escherichia coli. Arch Microbiol. 2008 doi: 10.1007/s00203-008-0406-2. [DOI] [PubMed] [Google Scholar]
- Harcum SW, Haddadin FT. Global transcriptome response of recombinant Escherichia coli to heat-shock and dual heat-shock recombinant protein production. J Ind Microbiol Biotechnol. 2006;33:801–814. doi: 10.1007/s10295-006-0122-3. [DOI] [PubMed] [Google Scholar]
- Kabir MM, Shimizu K. Gene expression patterns for metabolic pathway in pgi knockout Escherichia coli with and without phb genes based on RT-PCR. J Biotechnol. 2003;105:11–31. doi: 10.1016/S0168-1656(03)00170-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press, Cold Spring harbor, NY; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Five tables.
Global regulators and its regulated genes.