Abstract
A major goal of contemporary studies of embryonic development is to understand large sets of regulatory changes that accompany the phenomenon of embryonic induction. The highly resolved sea urchin pregastrular endomesoderm–gene regulatory network (EM-GRN) provides a unique framework to study the global regulatory interactions underlying endomesoderm induction. Vegetal micromeres of the sea urchin embryo constitute a classic endomesoderm signaling center, whose potential to induce archenteron formation from presumptive ectoderm was demonstrated almost a century ago. In this work, we ectopically activate the primary mesenchyme cell–GRN (PMC-GRN) that operates in micromere progeny by misexpressing the micromere determinant Pmar1 and identify the responding EM-GRN that is induced in animal blastomeres. Using localized loss-of -function analyses in conjunction with expression of endo16, the molecular definition of micromere-dependent endomesoderm specification, we show that the TGFβ cytokine, ActivinB, is an essential component of this induction in blastomeres that emit this signal, as well as in cells that respond to it. We report that normal pregastrular endomesoderm specification requires activation of the Pmar1-inducible subset of the EM-GRN by the same cytokine, strongly suggesting that early micromere-mediated endomesoderm specification, which regulates timely gastrulation in the sea urchin embryo, is also ActivinB dependent. This study unexpectedly uncovers the existence of an additional uncharacterized micromere signal to endomesoderm progenitors, significantly revising existing models. In one of the first network-level characterizations of an intercellular inductive phenomenon, we describe an important in vivo model of the requirement of ActivinB signaling in the earliest steps of embryonic endomesoderm progenitor specification.
Author Summary
In recent years, “gene regulatory networks” (GRNs) have provided integrated views of gene interactions that control biological processes. One of the earliest networks to be activated in the developing zygotes is the one controlling endomesoderm development. In the sea urchin, this network includes several subnetworks that function in adjacent tiers of cells that form the endoderm and mesoderm of the developing embryo. Although classic embryological manipulations have shown that the precursors of the embryonic skeleton induce endomesoderm fate in adjacent cells, the GRNs regulating this interaction are not understood. To investigate these networks, we ectopically activated a GRN that operates in skeletogenic precursors and characterized the responding GRN in neighboring cells, which adopt an endomesoderm fate. By testing the responsiveness of every core factor in the responding GRN, which allowed us to identify a subset that executes the response to the induction, we demonstrated that the signaling molecule, ActivinB, is an essential component of this induction and that its function is physiologically relevant: it is required during normal embryonic development to activate the same GRN that responds to signals from skeletogenic precursors. Furthermore, the network response to ActivinB signaling reveals greater complexity in an additional uncharacterized inductive signal emitted by skeletogenic precursors. Our results thus highlight how interacting GRNs can be used to understand a fundamental signaling process.
A classic embryonic induction in sea urchins is described in terms of interacting gene regulatory networks and a signal transduction system that connects them.
Introduction
Most animal embryos are patterned through a series of cell–cell interactions that shape and refine maternally inherited axial asymmetries. Cells are directed to assume specific fates through the phenomenon of embryonic induction, a process that has been intensely studied by developmental biologists for a number of decades. Understanding the mechanisms of embryonic induction is also of vital clinical importance, especially in the field of regenerative medicine, in which pluripotent stem cells are induced to differentiate into specific lineages of therapeutic interest. The widespread availability of sequenced eukaryotic genomes and microarray-based tools has greatly facilitated large-scale analyses of genes expressed at various times in the course of an embryonic induction. There is now a critical need to understand the regulatory interactions among these gene products that ultimately determine the outcome of the induction process.
By integrating regulatory interactions among large sets of gene products, gene regulatory networks (GRNs) provide uniquely global perspectives on biological and disease processes. Network-based approaches have therefore been adopted to describe Drosophila embryonic patterning [1], immune cell function [2], nematode vulval development [3], vertebrate retinal development [4], metabolic pathways [5], and malignant transformations [6]. The sea urchin endomesoderm–gene regulatory network (EM-GRN) was one of the first large-scale integrations of maternal and zygotic regulatory information in early embryonic development [7] and is currently one of the most detailed biological networks in existence [8]. A number of recent studies employing the sea urchin EM-GRN have demonstrated that, in addition to describing the regulatory blueprint underlying embryogenesis, changes in developmental GRN structure can suggest the molecular basis of the evolution of species-specific cell types and body plans across distant phyla [9,10].
An asymmetric localization of nuclear β-catenin is observed in most deuterostome embryos during early stages of development [11–15]. In sea urchin embryos, this anisotropy occurs along the primary (animal–vegetal) axis [13]. The vegetal wave of nuclearization of β-catenin during early cleavage stages is essential for endomesoderm formation, since embryos that lack nuclear β-catenin function develop as primarily ciliated ectoderm without any endomesodermal tissue [13]. These findings provided the initial experimental leverage for constructing the overall sea urchin pregastrular EM-GRN [7]. The constituent subnetworks of the overall EM-GRN include core sets of transcription factors that function cell autonomously and interact within specific blastomeres that eventually differentiate into primary (primary mesenchyme cell–GRN [PMC-GRN]) and secondary mesoderm (secondary mesoderm–GRN) as well as early endoderm (early endomesoderm/endoderm–GRN [E-EM/En-GRN] and late endoderm-GRN) derivatives (Figure 1A and [8]). Whereas regulatory interactions within each component network of the overall EM-GRN have been studied in considerable detail, critical connections between blastomeres, and thus between individual subnetworks, are not completely understood.
Figure 1. Gene Regulatory Networks Underlying Endomesoderm Induction.
(A) The micromere determinant Pmar1 (circled in red) activates the PMC-GRN in micromere progeny and is sufficient for micromere-derived endomesoderm-inducing signals. The E-EM/En-GRNs (up to 17 h postfertilization) integrate the regulatory functions of maternal and zygotic core factors that drive the earliest steps of endomesoderm progenitor specification in sea urchin embryos. The zygotically expressed core factors Z13, Eve, Wnt8, Blimp1, FoxA, and Brachyury (Bra) (circled in black) accumulate in presumptive endomesoderm during early developmental stages and could potentially respond to early inductive inputs from micromere descendants.
(B) Schematic depicting an experiment that reveals micromere-derived endomesoderm inductive signals, which are sufficient to induce ectopic endo16 expression and complete archenteron formation in animal blastomeres, and are also necessary for normal vegetal endo16 expression and timely gastrulation in the sea urchin embryo. The regulatory interactions among these signals and the overall EM-GRN are unknown. GRN diagram is adapted from [8].
One of the earliest inductive phenomena reported in experimental embryology was provided by the vegetal micromeres of the sea urchin embryo. Almost a century ago, Hörstadius reported that micromeres placed at the animal pole of the sea urchin embryo could induce adjacent presumptive ectoderm to differentiate as endodermal vesicles [16]. Several decades later, a molecular analysis confirmed that transplanted micromeres induce expression of the endomesoderm marker endo16 and eventually formation of a complete secondary archenteron from adjacent presumptive ectoderm (Figure 1B) [17]. Subsequent analyses have provided evidence in the normal embryo for a signal sent by micromeres and their progeny between fourth and sixth cleavages, which is required for normal endomesoderm specification, as monitored by endo16 expression, as well as for timely gastrulation (Figure 1B) [18]. Micromeres continue to emit archenteron-inducing signals through late cleavage/early blastula stages [19]. Despite several decades of efforts, however, neither the molecular identity(s) of these micromere-originated signals nor their specific regulatory contribution to early endomesoderm specification has been elucidated. Collectively, this constitutes a set of major unanswered questions in echinoderm embryo development.
The β-catenin–dependent determinant Pmar1 is transiently expressed in micromere progeny during the initial stages of specification of this cell lineage [20]. Pmar1 is sufficient for micromere-originated endomesoderm inducing signals and can regulate most aspects of micromere differentiation through activation of the PMC-GRN [21,22]. Pmar1′s ability to activate the PMC-GRN in any blastomere provides a unique opportunity to understand how signaling outputs of the PMC-GRN are interpreted through the regulatory logic of the recipient E-EM/En GRN (Figure 1A).
In this study, we resolve the regulatory connections between the Pmar1-activated PMC-GRN and the E-EM/En-GRN by testing the responsiveness of every zygotically activated core factor of the E-EM/En-GRN to the ectopic expression of the micromere determinant Pmar1. Our studies define a subset of the E-EM/En-GRN that responds to signals from the Pmar1-activated PMC-GRN and show that a significant fraction of the E-EM/En-GRN, including the recently identified Wnt8/Blimp1/β-catenin subcircuit [23], is insensitive to signals emitted by Pmar1-misexpressing blastomeres. We show that the transforming growth factor-β cytokine, ActivinB, is required both in Pmar1-expressing cells that emit endo16-inducing signals and in blastomeres that respond to this induction, making it an essential component of both emitting and responding GRNs. Furthermore, we demonstrate that ActivinB function is necessary for activation of the Pmar1-inducible subset of the E-EM/En-GRN in early endogenous endomesoderm development, whereas the Pmar1-unresponsive portion of the E-EM/En-GRN operates in vegetal endomesoderm precursors independently of ActivinB signaling. These key findings strongly suggest that ActivinB has an essential role in endomesoderm specification mediated by micromeres and their progeny and unexpectedly reveal the existence of a new micromere-derived signal(s). Collectively, we provide a critical global view of the regulatory interactions that constitute a classic embryonic induction response.
Results
Pmar1-Mediated Respecification of Ectoderm Is Regulated by a Subset of Core Factors within the Overall EM-GRN
Previous studies have shown that the micromere determinant Pmar1, through activation of the PMC-GRN, is sufficient for most aspects of micromere differentiation [20–22]. Pmar1 misexpression is sufficient to confer properties of the micromere/PMC lineage to all blastomeres in the embryo, including the ability to induce endo16 expression and formation of a complete ectopic gut from adjacent blastomeres [20,21]. Therefore, in order to determine which core factors of the E-EM/En-GRN respond to outputs of the PMC-GRN, we generated clones of cells expressing Pmar1 and GFP under the control of the hatching enzyme (SpHE) promoter (Figure 2A), which is active in nonmicromere lineages of the sea urchin embryo during early cleavage and blastula stages [24]. Consistent with previous studies [21], ectopic Pmar1-misexpressing cells ingressed into the blastocoel at the same time as endogenous primary mesenchyme cells. Furthermore, at the mesenchyme blastula stage, endo16 transcripts accumulated in ectopic nonvegetal blastomeres adjacent to gfp-positive, Pmar1-expressing cells, as well as in endogenous endomesoderm progenitors ([21]; see below). Collectively, these observations validate the use of the SpHE-pmar1 construct to examine ectopic E-EM/En-GRN activation in response to Pmar1-dependent induction signals.
Figure 2. EM-GRN Factors Induced by the Pmar1-Dependent Endomesoderm Induction.
(A) In all cases shown in (B–D), two-color FISH was used to detect ectopic induction of test transcripts (green) adjacent to gfp (red)-mRNA–expressing blastomeres in embryos injected with 0.02 μg/μl SpHE-gfp and 0.015 μg/μl SpHE-pmar1 (e–h in each case) compared to embryos injected with 0.02 μg/μl SpHE-gfp alone (a–d in each case).
Test transcripts: (B) z13 expression at 16–18 h p.f. (C) foxA expression at 20 h p.f.
(D) eve expression at 14 h p.f. Embryos shown are representative of over 80% of at least 50 embryos in each of three separate experiments in which gfp mRNA was detected in nonvegetal regions.
Black scale bar in (B) d represents approximately 20 μm. DIC images in (B) d and h, (C) d and h, and (D) d and h illustrate the absence of detectable developmental defects in injected embryos. gfp-expressing cells containing Pmar1 in (C) g and h have adopted a mesenchyme phenotype and ingressed into the blastocoel at 20 h p.f.
The E-EM/En-GRN includes the core regulatory factors, Z13 [25], Blimp1 [26], Wnt8 [27], Eve [28], FoxA [29], and Brachyury [30], all of which are zygotically expressed in endoderm and secondary mesoderm precursors during early cleavage and blastula stages (Figure 1A). We tested for transcripts encoding each of these core factors in cells adjacent to gfp-positive, Pmar1-misexpressing cells. At late cleavage and blastula stages, Pmar1 induced ectopic expression of z13 (Figure 2B, panels e–h vs. a–d), foxA (Figure 2C, panels e–h vs. a–d) and eve (Figure 2D, panels e–h vs. a–d) mRNAs within two to three cell diameters of gfp-labeled blastomeres. As for the late endomesoderm marker, endo16, each of these early genes was ectopically induced in presumptive ectoderm at the same developmental time that its transcripts accumulated in normal vegetal endomesoderm progenitors. In contrast to this set, no ectopic expression of blimp1 (Figure 3B, panels e–h vs. a–d), wnt8 (Figure 3C, panels e–h vs. a–d) or brachyury (Figure 3D, panels e–h vs. a–d) was detectable in cells adjacent to Pmar1-misexpressing clones at developmental times ranging from 10 to 22 h postfertilization (p.f.), when these transcripts accumulated in presumptive vegetal endomesoderm. These observations strongly suggest that Pmar1-misexpressing blastomeres respecify presumptive ectoderm as endomesoderm by activating only part of the E-EM/En-GRN, consisting of the genes encoding the core regulatory factors Z13, Eve, and FoxA. Ectopic activation of blimp1, wnt8, and brachyury was not detectable, thereby defining a significant part of the E-EM/En-GRN that is not activated by Pmar1-dependent induction.
Figure 3. EM-GRN Factors That Are Not Induced by Ectopic Pmar1 Expression.
(A) In all cases shown in (B–D), two-color FISH showed the absence of ectopic induction of test transcripts (green) adjacent to gfp-mRNA–expressing (red) blastomeres in embryos injected with 0.02 μg/μl SpHE-gfp and 0.015 μg/μl SpHE-pmar1 (e–h in each case) compared to embryos injected with 0.02 μg/μl SpHE-gfp (a–d in each case).
Test transcripts:
(B) blimp1 expression at 14–16 h p.f.
(C) wnt8 expression at 14–16 h p.f.
(D) brachyury expression at 20 h p.f.
Embryos shown are representative of 100% of at least 50 embryos in each of three separate experiments in which gfp mRNA was detected in nonvegetal regions. DIC images in (B ) d and h, (C) d and h, and (D) d illustrate the absence of detectable developmental defects in injected embryos. gfp-expressing cells containing Pmar1 in (D) panels g and h have adopted a mesenchyme phenotype and ingressed into the blastocoel at 20 h p.f.
ActivinB Is a Critical Component of Both PMC and EM-GRNs Underlying Pmar1- Dependent Induction
Members of the Nodal/Activin/TGFβ class of transforming growth factor-β cytokines have been implicated in endomesoderm development in vertebrate embryos [31], making them promising candidates for ligands that regulate micromere-dependent endomesoderm induction. The Strongylocentrotus purpuratus genome encodes several candidate TGFβ-related factors that are expressed before the hatching blastula stage [32]. Two of these, Nodal and Univin, are required for oral–aboral specification in the sea urchin embryo, but neither is necessary for endomesoderm formation [33,34]. In addition to these, transcripts encoding an ortholog of ActivinB (GenBank accession number: EU526314) were detected at low levels at developmental times (see below) when micromere progeny emit endomesoderm-inducing signals [18,19]. When ActivinB translation was blocked with either of two different antisense morpholino oligonucleotides (MO), gastrulation was consistently delayed by 12–18 h compared to buffer-injected controls (Figure 4A, panels d and g vs. a), and embryos contained supernumerary pigment cells at 3 d p.f. (Figure 4A, panels e and f, and h and i vs. b and c). Furthermore, we found that expression of endo16, the cardinal marker of micromere-mediated ectopic [17] and endogenous [18] endomesoderm induction, was significantly reduced in veg2 progeny at the mesenchyme blastula stage in ActivinB morphants (Figure 4C, panel c vs. a). We observed a similar gastrulation defect (Figure 4B, panels c and d vs. a and b) and inhibition of endo16 mRNA expression (Figure 4C, panel d vs. b) when we used the small molecule inhibitor, SB-431542 (SB; 5 μM) [35], to block signaling through Activin-like kinase-4/5/7 (ALK-4/5/7) during the first day of embryogenesis. Since ALK4/5/7 is a type I TGFβ receptor that transduces Activin/Nodal/TGFβ-like signals in the sea urchin embryo [34,36], these findings suggest that ActivinB signaling through the SB-sensitive ALK4/5/7 complex is necessary for normal archenteron formation in the sea urchin embryo and could constitute at least part of the signaling machinery required for endomesoderm induction in response to micromere signals.
Figure 4. ActivinB-ALK4/5/7 Function Is Required for Timely Gastrulation and Pregastrular endo16 mRNA Expression.
(A) Embryos were injected with buffer (Control; a–c) or 1.2 mM ActivinB MO1 (ActBMO1; d–f) or 0.6 mM ActivinB MO2 (ActBMO2; g–i) and photographed at 36 (a, d, and g) and 72 (b, c, e, f, h, and i) h p.f.
(B) Embryos were treated with DMSO (a and b) or 5 μM SB-431542 (c and d) for the first day of development and photographed at 36 (a and c) and 72 (b and d) h p.f. Compared to buffer-injected controls ([A] a), gut formation was delayed in embryos injected with ActivinB MO1 ([A] d) and ActivinB MO2 ([A] g). Similarly, gastrulation was delayed in SB-treated embryos ([B] c) compared to DMSO-treated controls ([B] a).
(C) Reduced endo16 mRNA expression was observed at 26 h p.f. in 1.2 mM ActBMO1-injected (c) and 5 μM SB-treated (d) embryos compared to buffer-injected (a) or DMSO-treated (b) controls respectively. Images are representative of the phenotypes observed in over 80% of at least 100 embryos in three separate experiments.
Black scale bar in (A) b represents approximately 40 μm.
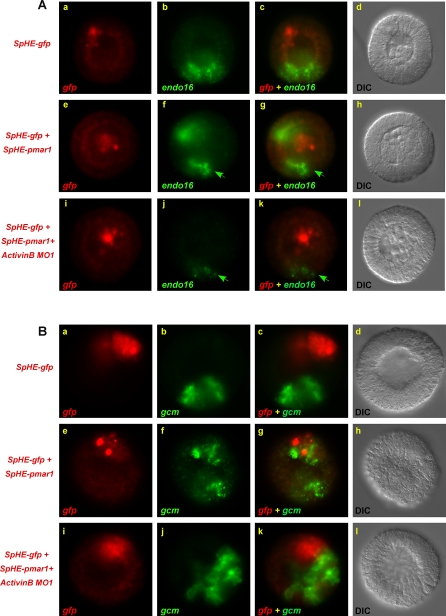
To determine whether ActivinB is necessary for ectopic endomesoderm induction, we assessed its requirement for endo16 induction adjacent to blastomeres expressing the micromere determinant Pmar1. Consistent with previous studies [21], in embryos injected with SpHE-gfp and SpHE-pmar1, endo16 mRNA accumulated at the mesenchyme blastula stage in ectopic nonvegetal patches of cells adjacent to Pmar1-expressing blastomeres, as well as in its normal vegetal territory (Figure 5A, panels e–h vs. a–d). When ActivinB MO was coinjected with the SpHE-gfp and SpHE-pmar1 constructs, both ectopic and endogenous endo16 expression were significantly reduced (Figure 5A, panels i–l vs. e–h). This finding identifies ActivinB as an essential component of Pmar1-dependent endomesoderm induction.
Figure 5. ActivinB Is Required for Pmar1-Mediated Endomesoderm Induction.
Embryos were injected with either 0.02 μg/μl SpHE-gfp or 0.02 μg/μl SpHE-gfp and 0.015 μg/μl SpHE-pmar1, as indicated to the left of the images. Embryos were analyzed by two-color FISH for gfp (red) and either (A) endo16 (green) at 26 h p.f. or (B) gcm (green) transcripts at 18–20 h p.f. The presence of 1.4 mM ActivinBMO1 strongly reduces both ectopic and endogenous endo16 mRNA levels ([A], i–l vs. e–h, compare arrowheads in [A] j and k vs. f and g for residual endogenous endo16 expression in the presence of ActivinMO1), but not those of the pigment cell marker, gcm ([B] i–l vs. e–h). Embryos shown are representative of more than 80% of a minimum of 50 embryos in each of three separate experiments in which gfp was detected in nonvegetal regions of the embryo. DIC images in (A) d and (B) d, h, and l illustrate the absence of detectable developmental defects in injected embryos. gfp-expressing cells containing Pmar1 in (A) h and l have adopted a mesenchyme phenotype and ingressed into the blastocoel at 26 h p.f.
Pmar1 is sufficient to activate all known inductive properties of micromeres, and cells misexpressing this determinant express Delta [20], which signals from micromeres through the Notch receptor to induce expression of the pigment cell marker, gcm, in neighboring blastomeres (Figure 5B, panels e–h vs. a–d) [37,38]. In contrast to endo16 induction, both endogenous and ectopic gcm induction were unaffected by coinjection of ActivinB MO along with the SpHE-gfp and SpHE-pmar1 constructs (Figure 5B, panels i–l vs. e–h). Thus, ActivinB is specifically required for Pmar1-mediated respecification of ectoderm as endomesoderm and not for micromere Delta-dependent secondary mesenchyme induction [37].
To test whether ActivinB function is required within Pmar1-expressing cells or within the cells that receive signals from them, we microinjected pmar1 and gfp mRNAs into one blastomere of a two-cell embryo and ActivinB MO into either the same blastomere or its sister. When ActivinB translation was inhibited exclusively within the Pmar1-misexpressing cells, induction of ectopic endo16 mRNA (at 26 h p.f.) in the other half of the embryo was significantly compromised (Figure 6, panels g–j vs. b–e). A small region of residual endogenous endo16 transcript accumulation persisted in the uninjected embryo half, demonstrating that the morpholino did not diffuse to the progeny of the uninjected blastomere (arrowheads in Figure 6, panels i and j). The descendants of the injected blastomere, on the other hand, adopted a primary mesenchyme cell fate and therefore did not express endo16. When ActivinB translation was blocked only in the recipient blastomere, both ectopic and endogenous endo16 mRNA expression were significantly down regulated in the progeny of that cell (Figure 6, panels k–o vs. a–e). These findings demonstrate that ActivinB is an essential component of the PMC-GRN that is activated in Pmar1-expressing blastomeres, as well as of the responding EM-GRN that operates in neighboring cells. This dual requirement suggests that ActivinB from Pmar1-expressing cells probably activates an autoregulatory loop in recipient cells that is necessary for amplifying its own response, as has been shown for other members of the Nodal/Activin subfamily of TGFβ cytokines [31]. Consistent with its function in both regulatory subnetworks, as well as the ability of all early blastomeres to respond to induction by micromeres, activinB transcripts accumulate throughout the embryo at early cleavage and blastula stages of embryogenesis (Figure 7, panels a–c) when micromere-derived endomesoderm inducing signals are sent [16,17,39]. Although Pmar1 induction of early endomesoderm specification requires ActivinB function, ectopic Pmar1 expression does not induce activinB mRNA expression (unpublished data). Additional studies will be required to determine the molecular basis of the interaction between Pmar1 and ActivinB.
Figure 6. ActivinB Is Required in Cells That Emit and Receive Pmar1-Dependent Induction Signals.
Individual blastomeres of embryos were injected as diagrammed to the left and analyzed by two-color FISH for gfp (red) and endo16 (green) transcripts. The concentrations of gfp mRNA, pmar1 mRNA, and ActivinB-MO1 used were 0.5 μg/μl, 0.2 μg/μl, and 1.4 mM, respectively. The presence of ActivinB MO1 in Pmar1-expressing cells (f–j) strongly inhibits ectopic and not endogenous (arrowheads in i and j) endo16 mRNA expression in the uninjected half of the embryo. When ActivinB MO1 is injected exclusively into blastomeres that respond to Pmar1-dependent induction (k–o), both ectopic and endogenous endo16 mRNA expression are strongly reduced. At the mesenchyme blastula stage shown, Pmar1 (gfp)-expressing cells in c, h, m and b, g, l acquire mesenchymal character and ingress into the blastocoel. Images represent the phenotypes observed in 16/19 (a–e), 43/47 (f–j), and 6/6 (k–o) injected embryos.
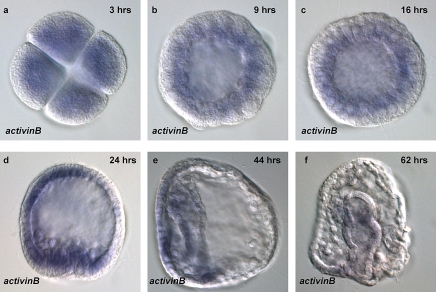
Figure 7. activinB mRNA Distribution during Early Sea Urchin Development at the Stages Indicated.
During early blastula and cleavage stages of development (a–c), low levels of activinB transcripts accumulate throughout the embryo. At the mesenchyme blastula stage (d), activinB mRNA is expressed in most of the embryo except one side of the ectoderm. At the late gastrula stage (e), activinB mRNA expression is detected in the gut and oral ectoderm and in the gut in prism (f) stage embryos.
ActivinB Signaling in Normal Endomesoderm Development Activates the Pmar1-Inducible E-EM/En-GRN Response
The data described thus far resolve a section of the E-EM/En-GRNs that is activated in response to Pmar1-derived endomesoderm-inducing signals. Furthermore, we demonstrate that Pmar1-dependent ectopic induction of the cardinal endomesoderm marker, endo16, requires ActivinB. Previous studies have shown that micromere-derived signal(s) regulate endo16 accumulation, not only in presumptive ectoderm, but also in the endogenous vegetal plate [17,18]. Therefore, our finding that ActivinB is required for endo16 accumulation in both ectopically induced and endogenous locations strongly suggests that it mediates micromere-dependent endomesoderm specification in the vegetal plate. Furthermore, as in micromereless embryos, gastrulation is significantly delayed in ActivinB morphants and in embryos treated with SB, which inhibits ALK4/5/7 function [35,36]. ALK4/5/7 function in promoting timely gastrulation is required from early cleavage to blastula stages when micromere progeny signal to overlying veg2 macromere descendants [19] because pulses of 5 μM SB for at least a 4-h interval any time prior to hatching at 18 h p.f. delay gastrulation by 12–18 h, whereas treatment after 18 h p.f. had no effect on the timing of gut development (unpublished data). These results are consistent with a role for ActivinB signaling in early endoderm development.
To determine whether the Pmar1-responsive EM-GRN resolved by the above experiments accurately describes the GRN response to ActivinB signaling in vegetal blastomeres during normal sea urchin development, we characterized the requirement for ALK4/5/7-ActivinB function in pregastrular endomesoderm specification. Whole-mount in situ hybridizations showed that SB treatment significantly inhibits the accumulation of transcripts encoding the core regulatory factors, Z13 (Figure 8A, panel g vs. a), Eve (Figure 8A, panel h vs. b), and FoxA (Figure 8A, panel i vs. c) in the veg2 tier of blastomeres (see fate map in Figure 8C) prior to the hatching blastula stage. The reduction in z13 and eve expression in drug-treated embryos was observed as early as 8 h (∼seventh cleavage) following fertilization. Similarly, a MO-mediated block of ActivinB translation strongly inhibited the expression of z13 (Figure 8A, panel j vs. d) and foxA (Figure 8A, panel l vs. f) mRNAs when assayed at the corresponding stages. We did not detect any difference in the expression of eve mRNA in veg2 blastomeres in ActivinB morphants at cleavage stages (Figure 8A, panel k vs. e), suggesting that this transcription factor may be regulated by a TGFβ cytokine other than ActivinB. In contrast, the accumulation of transcripts encoding the remaining members of the E-EM/En-subnetworks, brachyury, blimp1, and wnt8, did not depend on ALK4/5/7 (Figure 8B, panels g, h, and i vs. a, b, and c, respectively) or ActivinB (Figure 8B, panels j, k, and l vs. d, e, and f, respectively) function.
Figure 8. ActivinB-ALK4/5/7 Signaling Activates the Pmar1-Responsive E-EM/En-GRN during Normal Endomesoderm Specification.
(A) Embryos were treated with either (a–c) DMSO (Control) or (g–i) SB-431542 (SB; 5 μM) and analyzed by WMISH to detect (a and g) z13 at 8 h , (b and h) eve at 8 h, (c and i) foxA at 18 h. SB treatment strongly inhibits the accumulation of each of these transcripts in veg2 blastomeres at the stages assayed (g–i) compared to DMSO-treated controls (a–c). The same mRNAs were detected in embryos injected either with buffer (d–f) or ActivinB MO1 (ActBMO1; 1. 2 mM) (j–l) at 14 h (d and j), 10 h (e and k), or 20 h (f and l), respectively. z13 and foxA mRNA expression in veg2 blastomeres was strongly reduced in ActivinB MO1-injected embryos (j and l) compared to buffer-injected controls (d and f) assayed at corresponding stages. At the late cleavage stage, eve mRNA expression in the veg2 tier was unaffected by the presence of ActivinB MO1 (k vs. e).
(B) Embryos were treated with either (a–c) DMSO or (g–i) SB (5 μM) and analyzed by WMISH to detect (a and g) brachyury at 18 h, (b and h) blimp1 at 14 h, and (c and i) wnt8 at 14 h. The same mRNAs were detected in embryos injected either with buffer (d–f) or ActBMO1 (1.2 mM) (j–l) at 20 h (d and j), 14 h (e and k), and 14 h (f and l), respectively. The accumulation of each of these transcripts in veg2 blastomeres was independent of ALK4/5/7 (g–i vs. a–c) and ActivinB (j–l vs. d–f) function at the stages assayed.
Images in (A) and (B) represent the molecular phenotypes seen in over 80% of at least 50 embryos in three separate experiments.
(C) Fate map of blastula stage embryo.
Pigment cells (a secondary mesoderm derivative) were formed at similar times in controls and in embryos lacking ALK4/5/7 or ActivinB function. Consistent with this observation, we found that the first phase of Delta-Notch–mediated secondary mesoderm specification, as assayed by expression of gcm mRNA in veg2 progeny [37,38] was independent of ActivinB-ALK4/5/7 activity (Figure 9A, panel e vs. a and panel g vs. c). Similarly, gataE was expressed normally in veg2 secondary mesoderm precursors of SB-treated embryos (Figure 9A, panel f vs. b) as well as ActivinB morphants (Figure 9A, panel h vs. d). However, at 34 h p.f., ActivinB morphants contained 40% more gcm-positive pigment cell precursors than did buffer-injected controls (Figure 9B, panels d–f vs. a–c), probably due to suppression of early FoxA expression, which represses gcm transcription in veg2 endoderm precursors [29]. Thus, similar to its regulatory properties in ectopic Pmar1-mediated induction, normal vegetal activation of the secondary mesoderm (pigment cell)-GRN does not require ActivinB or ALK4/5/7 function.
Figure 9. Secondary Mesoderm Specification Is Independent of ActivinB-ALK4/5/7 Function.
(A) Embryos were treated with either (a and b) DMSO (Control) or (e and f) SB-431542 (5 μM) and analyzed by WMISH to detect (a and e) gcm at 18 h and (b and f) gataE at 18 h. The same mRNAs were detected in embryos injected either with (c and d) buffer or (g and h) ActivinB MO1 (1.2 mM) at 18 h (c, g, d, and h). The accumulation of each of these transcripts in veg2 secondary mesoderm precursors was independent of ALK4/5/7 (e and f vs. a and b) and ActivinB (g and h vs. c and d) function at the stages assayed. All images are representative of the molecular phenotypes observed in more than 80% of a minimum of 50 embryos in each of three separate experiments.
(B) At 34 h p.f., (a–c) buffer-injected controls contain 29 gcm-mRNA expressing (n = 5 embryos examined) cells (green in b and c) compared to 40 (n = 5 embryos examined) such cells (green in e and f) in embryos injected with (d–f) 1.2 mM ActivinB MO1. Nuclei are stained with DAPI (blue).
By the late mesenchyme blastula stage, z13 and foxA transcripts accumulate in veg1 derivatives in addition to veg2 descendants (see fate map in Figure 10B) and the corresponding proteins are essential elements of the late endoderm-GRN. Unlike their earlier expression in the veg2 tier, this later phase of expression does not depend detectably on ALK4/5/7 or ActivinB function (Figure 10A, panel e vs. a, f vs. b, g vs. c, and h vs. d). This finding is consistent with the fact that SB treatment after hatching does not hinder gastrulation and demonstrates that ActivinB signaling through the SB-sensitive ALK4/5/7 complex regulates core elements exclusively within the E-EM/En-GRNs, which function in veg2 blastomeres overlying the micromeres.
Figure 10. The Late Endoderm-GRN Is Independent of ActivinB-ALK4/5/7 Signaling.
(A) Embryos were treated with either (a and b) DMSO (Control) or (e and f) SB-431542 (5 μM) and analyzed by WMISH to detect (a and e) z13 at 26 h and (b and f) foxA at 26 h . The same mRNAs were detected in embryos injected either with (c and d) buffer or (g and h) ActBMO1 (1.2 mM) at 26 h (c, g, d, and h). The late accumulation of each of these transcripts in vegetal blastomeres was independent of ALK4/5/7 (e and f vs. a and b) and ActivinB (g and h vs. c and d) function at the stages assayed. Images are representative of the molecular phenotypes observed in more than 80% of a minimum of 50 embryos in each of three separate experiments.
(B) Fate map of blastula-mesenchyme blastula stage embryo.
The findings described above demonstrate that the gastrulation defect observed in SB-treated embryos and ActivinB morphants can be entirely attributed to compromised early specification of veg2 endomesoderm progenitors. Interestingly, the GRN response to ActivinB-ALK4/5/7 signaling in normal pregastrular endomesoderm development is almost identical to that of ectopic Pmar1-driven induction. Collectively, these observations strongly suggest that a micromere-derived, ActivinB-dependent signal is necessary for early veg2 endomesoderm progenitor specification through the outputs of a discrete set of core factors of the E-EM/En subnetworks.
Veg2 Endomesoderm Progenitors Receive Multiple Signals from Micromere Progeny during Early Cleavage
Previous studies have shown that micromeres emit signals during early cleavage stages that clear the β-catenin antagonist SoxB1 from veg2 macromere progeny, a process that has been suggested to be important for the activation of the E-EM/En-subnetworks [21,27,40]. Similarly, when transplanted to animal locations, micromeres also clear SoxB1 protein from surrounding ectoderm cells during their respecification as endomesoderm [21]. Pmar1 misexpression is sufficient to induce SoxB1 down-regulation in adjacent blastomeres in both animal and vegetal contexts [21], and it has been proposed that this clearance occurs in response to the same Pmar1-dependent endomesoderm induction signal that activates endo16 expression in adjacent blastomeres [21,27]. Since ActivinB is an essential component of this Pmar1-derived induction signal and regulates both ectopic and endogenous endo16 accumulation, we examined its role in clearing SoxB1 from veg2 secondary mesoderm progenitors. If SoxB1 clearance requires ActivinB, then SoxB1 should persist at high levels in secondary mesenchyme of ActivinB morphants. Surprisingly, however, SoxB1 clears normally from this region, which is marked by gcm mRNA accumulation (Figure 11A, panels d–f vs. a–c). We conclude that ActivinB and an unidentified SoxB1-clearing signal are separate, early outputs of micromeres.
Figure 11. Pmar1-Responsive Endomesoderm Specification Is Independent of the Micromere Signal That Clears Vegetal SoxB1.
(A) SoxB1 protein (green), gcm mRNA (red), and DAPI nuclear staining (blue) at 18 h p.f.: (a–c) buffer-injected controls; (d–f) 1.2 mM ActivinB MO1–injected embryos; and (g–i) 0.15 mM Delta MO-injected embryos. Images represent the phenotype observed in over 80% of at least 50 embryos in each of three separate experiments. SoxB1 protein clears normally from veg2 secondary mesoderm precursors in ActivinB (d–f vs. a–c) and Delta (g–i vs. a–c) morphant embryos.
(B) (a–c) SoxB1 protein (green), z13 mRNA (red), and DAPI nuclear staining (blue) at 8 h p.f. in normal embryos showing high levels of SoxB1 protein in nuclei of z13-expressing cells at the cleavage stage of development. The optical section is slightly oblique with respect to the animal-vegetal axis, and z13-expressing cells are at the bottom.
The specific role of SoxB1 clearance in the EM-GRN remains to be elucidated. Although SoxB1 clearance is indicative of micromere signaling, it is gradual and not completed in veg2 progeny until the early mesenchyme blastula stage, after initial veg2 specification has occurred [41,42]. Consistent with this observation, we found that at cleavage stages, genes encoding the Pmar1-responsive endomesoderm core factors, z13 and eve, are expressed in blastomeres that contain high levels of SoxB1 (z13 shown in Figure 11B; eve data not shown), confirming that micromere-mediated early veg2 endomesoderm specification does not require SoxB1 clearance. This is also the case in presumptive endoderm since endoderm-specific genes are expressed normally when the later phase of micromere-dependent SoxB1 clearance is prevented [40]. Furthermore, although endogenous SoxB1 limits nuclear β-catenin activity and SoxB1 misexpression strongly antagonizes it, SoxB1 morphants have greatly reduced endoderm and fail to gastrulate [42], demonstrating that some level of SoxB1 is required for normal endoderm development. To test whether down-regulation of SoxB1 might instead be a consequence of early specification, we blocked Delta-mediated secondary mesoderm specification by use of a Delta-MO. This approach inhibited gcm expression, as expected [37,38], but had no effect on the extent of SoxB1 clearance from veg2 blastomeres (Figure 11A, panels g–i vs. a–c). Thus, micromere-dependent SoxB1 clearance occurs normally when at least two Pmar1-mediated veg2 specification signals (ActivinB and Delta) are blocked. We conclude that the signal for SoxB1 down-regulation, ActivinB, and Delta make largely separate Pmar1-dependent contributions to the development of endomesodermal tissues within the veg2 tier of blastomeres.
Discussion
This study describes one of the first systematic demonstrations of the developmental GRNs activated in blastomeres that emit and respond to embryonic endomesoderm induction signals. We ectopically activated the Pmar1-dependent PMC-GRN in nonvegetal cells and demonstrate that adjacent blastomeres are specified as endomesoderm through a responding GRN that includes genes encoding the core factors Z13, Eve, and FoxA. In contrast, a significant part of the sea urchin EM-GRN, consisting of the regulatory outputs of Wnt8, Blimp1, and Brachyury, is not activated through Pmar1-mediated signals. Two members of this Pmar1-insensitive set, Wnt8 and Blimp1, have recently been proposed to constitute a self-sustaining cis-regulatory subcircuit that maintains nuclearization of β-catenin in endomesoderm progenitors [23]. Since ectopic Pmar1-misexpressing cells do not detectably induce wnt8 or blimp1 expression in adjacent animal blastomeres, our work implies that these responding cells are specified as endomesoderm without accumulating β-catenin in their nuclei. This conclusion is consistent with previous studies demonstrating the absence of detectable nuclear β-catenin in animal blastomeres specified as endomesoderm through ectopic micromere induction signals [13].
Using endo16 expression, the molecular hallmark of micromere-mediated endomesoderm specification [16,17,43], we show through localized loss-of-function analyses that the TGF-β cytokine, ActivinB, is required in both Pmar1-expressing cells that send endomesoderm-inducing signals as well as in the responding EM-GRN in blastomeres that transduce this signal. The requirement for ActivinB in Pmar1-dependent endomesoderm induction strongly suggests that it plays a similar essential role in endomesoderm formation in response to signals from ectopic micromeres, a phenomenon first described almost 70 y ago in the sea urchin embryo [16]. ActivinB signaling through ALK4/5/7 during early cleavage and blastula stages also is necessary for endogenous vegetal activation of almost the entire E-EM/En-GRN induced by ectopic Pmar1 expression. This critical finding, along with the fact that, like micromere progeny, ActivinB is required for normal endo16 expression in veg2 endomesoderm progenitors, and for timely gastrulation, strongly suggests that this TGFβ ligand plays an essential role in early micromere-dependent endomesoderm specification.
In contrast to the Pmar1-dependent EM-GRN factors, ActivinB is not required for normal vegetal expression of any of the Pmar1-insensitive EM-GRN core factors. Furthermore, both the Delta-dependent secondary mesoderm–GRN and the late endoderm-GRN (17 to 30 h p.f.) that is subsequently activated in veg2 and veg1 macromere progeny are independent of ActivinB function. Thus, the primary cellular targets of ActivinB signaling during pregastrular endomesoderm development are the more vegetal (veg2) macromere derivatives. The exact state of specification of these cells in ActivinB morphants is not clear, but some of them transfate to pigment cells, presumably as a result of reduced FoxA activity [29]. Whether any of these cells adopts other mesodermal fates is not yet known. If indeed they do not contribute to definitive endoderm, then the gut that eventually forms would arise from veg1 progeny that are specified much later than veg2 derivatives [44], through the regulatory outputs of the ActivinB-independent late endoderm-GRN. Alternatively, if veg2 endoderm progenitors in ActivinB morphants do not acquire other secondary mesoderm fates, then they probably participate in gut formation. In this case, either they eventually express all of the core EM-GRN factors through micromere-independent mechanisms or, in a later vegetal developmental context, the Wnt8/Blimp1/nuclear β-catenin subcircuit [23] is sufficient to support this process. Either scenario would require considerable regulatory cross talk between GRN components in order to achieve threshold concentrations of critical core factors that drive definitive endoderm development.
Recent models of pregastrular development in echinoderm embryos have proposed that a single micromere signal specifies veg2 macromere progeny as endo16-expressing endomesoderm progenitors and also causes the gradual clearance of the β-catenin antagonist, SoxB1, from nuclei of these blastomeres [21,27,40]. This is an attractive model because it invokes a causal linkage between activation and maintenance of the EM-GRN that depends on nuclear β-catenin accumulation and removal of an antagonist. However, the results of several experiments presented here argue strongly that micromere-mediated early endomesoderm induction and micromere-regulated SoxB1 removal are independent processes. First, SoxB1 down-regulation occurs normally in embryos lacking the early endomesoderm-inducing (endo16-inducing) signal ActivinB. Second, SoxB1 clearance is not required for activation of the early EM-GRN because macromere progeny express early Pmar1-responsive core factors in this subnetwork before SoxB1 protein levels are detectably reduced in nuclei of the same cells. Third, ectopic micromere-mediated EM-GRN activation and complete archenteron formation occur in cells that lack detectable nuclear β-catenin [13] but still clear SoxB1 [21]. Our findings are consistent with other studies showing that endoderm specification can occur without SoxB1 clearance [40]. The first phase of SoxB1 clearance in presumptive secondary mesoderm also does not depend on other known micromere signals, because it occurs normally in either Delta (this work) or Wnt8 morphants [27]. Therefore, although SoxB1 clearance depends on an early micromere signal(s), it is a gradual process that is not completed until the mesenchyme blastula stage and relies on an unknown pathway(s) distinct from the one that regulates endo16 induction through ActivinB (Figure 12). Therefore, we favor the view that fine-scale patterning of endoderm and mesoderm in different blastomere tiers probably requires micromere-dependent regulation of the level and duration of SoxB1 expression, but early micromere-mediated endomesoderm specification is independent of this process (Figure 12).
Figure 12. Pregastrular EM-GRN Subnetworks Mediating Micromere-Mediated Endomesoderm Induction.
The micromere determinant Pmar1 activates the PMC-GRN and transmits endomesoderm-inducing signals to macromere descendants. The Pmar1-responsive genes z13, foxA, eve (and the cardinal endomesoderm marker, endo16) define a responding EM-GRN (red arrows leading to factors circled with solid red lines) that is activated in animal blastomeres. The Pmar1-unresponsive genes wnt8, blimp1, and brachyury (Bra) (circled with hatched black lines) are dispensable for ectopic Pmar1-mediated endomesoderm induction, which probably occurs without inducing ectopic nuclearization of β-catenin in animal blastomeres. In both animal and vegetal contexts, ActivinB is necessary for Pmar1-activated endomesoderm induction and eventual endo16 expression (shown at bottom right of diagram) and is an essential component of the PMC-GRN and E-EM/En-GRNs. ES refers to an early signal, thought to depend on Pmar1, that is sent from micromeres starting after fourth cleavage stage, which, like ActivinB, is required for endo16 expression and timely gastrulation. ActivinB-independent Pmar1-derived Delta signaling (blue arrow) specifies pigment and blastocoelar cell fate within secondary mesoderm precursors and regulates gcm expression in veg2 descendants (the SMC-GRN core factor, Gcm, shown in the diagram is not an element of the E-EM/En-GRNs). A third, unknown Pmar1-mediated signal clears SoxB1 (black line leading to factor circled with solid black line) from endomesoderm precursors and, contrary to previous models of pregastrular endomesoderm development, is independent of micromere-mediated specification of these blastomeres. GRN diagram is adapted from [8].
This work is the first report, to our knowledge, of the requirement of ActivinB in the earliest steps of endomesoderm specification in any developmental model system and of the involvement of a TGFβ cytokine of the Activin/Nodal/TGFβ class in endomesoderm formation in an invertebrate embryo. Activin was initially proposed to be a mesoderm-inducing agent because exogenous Activin can induce mesoderm in amphibian animal cap explants [45]. Activin can also direct endoderm differentiation in human and mouse embryonic stem cells, which, interestingly, transit through an endomesoderm-like primordium prior to expressing markers of definitive endoderm [46,47]. Recent studies in which ActivinB was knocked down with MOs in amphibian embryos suggest that it does play some role in axial mesoderm formation, possibly by regulating convergent extension movements of gastrulation through the activities of other mesoderm-inducing factors [48] and/or the timing of cell cycle transitions in involuting dorsal axial mesoderm [49]. However, even though ActivinB is expressed during early development of amphibian embryos [50], there is, at present, no evidence suggesting that it functions in early endomesoderm specification per se. By analyzing the EM-GRN response to ActivinB, we show that it is exclusively required for the earliest steps of endomesoderm specification, eventually leading to the formation of definitive endoderm and timely gastrulation in the sea urchin embryo. This work, therefore, provides a critical in vivo model of the requirement of ActivinB function in early embryogenesis. Furthermore, the remarkable developmental plasticity of the sea urchin embryo allows us to demonstrate that ActivinB is the cardinal primary mesenchyme-derived signal that can activate an ectopic endomesoderm GRN. We thus describe an important experimental paradigm that elucidates the GRNs that drive endogenous and ectopic endomesoderm induction.
By defining major regulatory connections between individual networks of the overall sea urchin EM-GRN, we have built a new framework for future studies of endomesoderm specification. In the sea urchin embryo, endomesoderm development is highly regulative and eventually occurs even in the absence of micromeres [18,43]. Similarly isolated macromeres also express endoderm markers [51]. Our identification of a Pmar1-insensitive set of core regulatory factors may significantly inform future work addressing how the network responds in these situations, potentially leading to an understanding of the molecular basis of such regulative endomesoderm development. Our work also postulates the existence of an uncharacterized micromere signal that is emitted during early cleavage stages to clear the β-catenin antagonist SoxB1 from endomesoderm precursors. Identifying this signal and its GRN properties will be necessary to achieve a comprehensive understanding of pregastrular development in echinoderm embryos. We show that endomesoderm is ectopically induced through the regulatory outputs of a set of Pmar1-responsive EM-GRN core factors, making it important to understand how this core set activates and stabilizes downstream GRN circuits. Since this Pmar1-responsive GRN potentially drives formation of a complete archenteron without detectable nuclear β-catenin, it is also conceivable that additional uncharacterized β-catenin–independent GRNs exist. Interestingly, recent studies have demonstrated the existence of such a β-catenin-independent JNK/Axin dorsalization pathway in zebrafish embryos (e.g., [52]). The sea urchin embryo, with its well-known developmental plasticity, would be an ideal system for characterizing such nascent GRNs and examining their interactions with the currently understood endomesoderm regulatory networks.
Materials and Methods
Embryo cultures and inhibitor treatments.
Adult sea urchins (S. purpuratus) were obtained from Marinus Scientific (Garden Grove, California) or The Cultured Abalone (Goleta, California). After removal of fertilization envelopes, embryos were cultured at 15 °C in artificial seawater (ASW) at fewer than 300 embryos/ml of ASW. A stock solution of the ALK4/5/7 inhibitor SB-431542 (Tocris Biosciences) in dimethylsulfoxide was added to cultures to a final concentration of 5 μM. Inhibitor treatments were terminated by pipetting embryos through five ASW-containing 60 × 15-mm Petri dishes. Only cultures in which more than 90% control embryos were morphologically normal were used.
Microinjections.
Fertilized S. purpuratus eggs in 60 × 15-mm Petri dishes coated with 1% (w/v) protamine sulfate were microinjected using an Eppendorf Femtojet-Injectman NI2 micromanipulator attached to a Leica inverted microscope. Microinjections were performed in PABA-ASW followed by three rinses in ASW. Injection solutions containing 25% glycerol and 10 mM Tris-HCl (pH 8.0) were filtered through 0.22 μM PVDF filters (Millipore) and briefly incubated at 50 °C before loading to minimize clogging within the injection needles (Femtotips I; Eppendorf). For single-blastomere injections, embryos were cultured in ASW until 40 min after the first cell division and rinsed four times with ice-cold calcium-magnesium–free sea water (CMFSW). Embryos were microinjected within 10 min of the last CMFSW rinse, washed four times with ice-cold ASW, and cultured to the desired developmental stage at 15 °C.
Construct preparation, RNA synthesis, 5′ RACE, and morpholino sequences.
The SpHE-GFP construct contains the SpHE minimal promoter sequence (from −310 to +96, [24]) inserted between the EcoRI and SalI sites of pGreenLantern (pGL). For the SpHE-pmar1 construct, the GFP fragment from SpHE-GFP was replaced with the Pmar1 coding region (GenBank accession number: AF443277) and the first six base pairs upstream of its initiation codon. Both constructs were linearized with PstI prior to microinjection.
pmar1 and gfp mRNAs were synthesized from linearized pCS2+-pmar1 and pCS2+-gfp templates using the mMessage mMachine kit (Ambion). RNA concentrations were measured by spectrophotometry, and RNA integrity was verified by electrophoresis through agarose-formaldehyde gels prior to microinjection.
To determine the 5′-end of activinB mRNA, RACE (rapid amplification of cDNA ends)-ready cDNA was synthesized from 1 μg of DNA-free total RNA extracted from eggs using the SMART RACE cDNA synthesis kit (BD Biosciences) per the manufacturer's instructions. A touchdown-PCR protocol recommended by the manufacturer was used to amplify an activinB 5′ RACE product with the activinB gene–specific reverse primer, (5′-TCACGAACACCCACAACTCAAGATCCTCATTTC-3′) and the kit's SMART oligonucleotide forward primer mix. RACE products were cloned into PGEM T-Easy (Promega), and 20 clones were sequenced completely in both directions to determine the 5′-end of activinB. The activinB mRNA sequence has been deposited in GenBank with the accession number of EU526314.
Morpholino-substituted antisense oligonucleotide (MO) sequences.
Sequences were as follows: SpActivinB MO1: 5′-gcaggtacagagcttctcggtcaac-3′; SpActivinB MO2: 5′-TATCAAGCTGGCCCACGACATGAAA-3′; and Delta MO: 5′-GCCGATCCGTTATTCCTTTCTTATC-3′.
MOs were dissolved in 100 μl of nuclease-free water to give 3 mM stock solutions, which were diluted to the concentrations indicated in the figure legends.
Whole-mount in situ hybridizations (WMISH) and immunostains.
Embryos were fixed in 4% paraformaldehyde-ASW containing 10 mM MOPS (pH 7.0) and 0.1% Tween-20 for 1 h at room temperature (RT) in 96-well flat-bottomed plates. Hybridization and detection procedures using colorimetric alkaline phosphatase-conjugated digoxigenin antibodies were carried out as described previously [53].
For single-color fluorescent in situ hybridizations (FISH) using a digoxigenin-conjugated RNA probe and tyramide signal amplification (TSA-Plus kit; PerkinElmer), the above procedure was modified as follows. After posthybridization washes, embryos were incubated for 1 h at RT in a blocking buffer consisting of 10% normal goat serum and 5 mg BSA/ml in MOPS wash buffer [53], followed by 30 min at RT in horseradish peroxidase (POD)-conjugated anti-digoxigenin antibody (Roche) (1:1,500) in blocking buffer. Embryos were washed eight times over the course of 2 h at RT with MOPS wash buffer. Hybrids were detected with a 1:100 dilution of TSA-stock solution (FITC TSA-Plus kit or Cy3-TSA Plus kit; PerkinElmer) in diluting buffer at RT for 6–8 min, followed by five washes in MOPS wash buffer at RT.
For dual-color FISH with fluorescein- and digoxigenin-conjugated RNA probes, after the blocking step, embryos were incubated in POD-conjugated anti-fluorescein antibody (Roche) (1:750) at RT overnight, followed by eight washes with MOPS wash buffer. Duplexes were detected by incubating embryos in Cy3 TSA (1:100 dilution) for 6–8 min, followed by five washes with MOPS wash buffer. The POD activity of the bound anti-fluorescein antibody was quenched with 0.1% (w/v) sodium azide in MOPS wash buffer for 30 min at RT, followed by six washes with MOPS wash buffer for a total of 60 min. Digoxigenin-labeled transcripts were detected subsequently using FITC-TSA as described above.
The dual-labeled FISH procedure was modified to allow hybridization of probes to endogenous transcripts and mRNA synthesized from injected SpHE promoter-containing plasmids but to exclude formation of hybrids with plasmid DNA [54]. To prevent DNA denaturation during hybridization, the formamide concentration was reduced to 50% and the hybridization temperature lowered to 45 °C. After 4–7 d of hybridization, unhybridized probe was removed with five washes in MOPS wash buffer at 50 °C for 3 h [54]. Residual binding of the probe to DNA was eliminated by two washes in standard hybridization buffer (70% formamide) for 30 min at 50 °C.
For single-color FISH and SoxB1 immunostaining double labeling, FISH was performed first as described above. Embryos were incubated overnight at 4 °C with SoxB1 primary antibody that had been filtered by centrifugation through a 0.22-μm PVDF mesh (Millipore) and diluted 1,000-fold in a buffer containing 5% normal lamb serum, 1× phosphate-buffered saline (PBS), and 0.05% Tween-20 (PBST blocking buffer). After five washes for a total of 1 h at RT with PBST wash buffer (1× PBS, 0.05% Tween-20), bound primary antibody was detected with goat anti-rabbit Alexa-488 secondary antibody (1:750; Invitrogen) in PBST blocking buffer for 1 h at RT.
Image processing.
Images were captured on a Zeiss Axiovert200 inverted microscope equipped with an Axiotome axial tomography device and differential interference contrast (DIC) optics in Axiovision Release 4.6. Images were processed in Adobe Photoshop 8.0 and Canvas X. Z-stack composites were generated using Imaris Release 5.7.2 (Bitplane).
Acknowledgments
We thank Diane Adams for comments on this manuscript, and Junko Yaguchi, Laurel Newman, Shunsuke Yaguchi, Zheng Wei, John Paul Masly, and Amy Sutton for helpful discussions.
Abbreviations
- ALK
Activin-like kinase
- E-EM/En-GRN
early endomesoderm/endoderm–gene regulatory network
- EM-GRN
endomesoderm–gene regulatory network
- FISH
fluorescent in situ hybridization
- GRN
gene regulatory network
- MO
morpholino oligonucleotide
- p.f.
postfertilization
- PMC-GRN
primary mesenchyme cell–gene regulatory network
- SB
SB-431542
- WMISH
whole-mount in situ hybridization
Footnotes
Author contributions. AJS conceived and designed the experiments and performed the experiments. AJS, RCA, and LMA analyzed the data. AJS and LMA contributed reagents/materials/analysis tools. AJS, RCA, and LMA wrote the paper.
Funding. This work was supported initially by a grant to RCA (NIH-GM25553) and recently by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests. The authors have declared that no competing interests exist.
References
- Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Singh H, Medina KL, Pongubala JMR. Contingent gene regulatory networks and B cell fate specification. Proc Natl Acad Sci U S A. 2005;102:4949–4953. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Wang M, Ririe TO, Fernandes JS, Sternberg PW. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc Natl Acad Sci U S A. 2005;102:4972–4977. doi: 10.1073/pnas.0408122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Beremand PD, Thomas TL, et al. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev Biol. 2005;280:467–481. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Chin C-S, Chubukov V, Jolly ER, DeRisi J, Li H. Dynamics and design principles of a basic regulatory architecture controlling metabolic pathways. PLoS Biol. 2008;6:e146. doi: 10.1371/journal.pbio.0060146. doi: 10.1371/journal.pbio.0060146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdollahi A, Schwager C, Kleeff J, Esposito I, Domhan S, et al. Transcriptional network governing the angiogenic switch in human pancreatic cancer. Proc Natl Acad Sci U S A. 2007;104:12890–12895. doi: 10.1073/pnas.0705505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, et al. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- de-Leon SB-T, Davidson EH. Gene regulation: gene control network in development. Annu Rev Biophys Biomol Struct. 2007;36:191–212. doi: 10.1146/annurev.biophys.35.040405.102002. [DOI] [PubMed] [Google Scholar]
- Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc Natl Acad Sci U S A. 2008;105:6091–6096. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman V, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci U S A. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Steinbeisser H, Warga RM, Hausen P. β-catenin translocation into nuclei demarcates the dorsalizing centers in frog and fish embryos. Mech Dev. 1996;57:191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- Rowning BA, Wells J, Wu M, Gerhart JC, Moon RT, et al. Microtubule-mediated transport of organelles and localization of β-catenin to the future dorsal side of Xenopus eggs. Proc Natl Acad Sci U S A. 1997;94:1224–1229. doi: 10.1073/pnas.94.4.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, Mcclay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- Roeser T, Stein S, Kessel M. Nuclear beta-catenin and the development of bilateral symmetry in normal and LiCl-exposed chick embryos. Development. 1999;126:2955–2965. doi: 10.1242/dev.126.13.2955. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, et al. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–450. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- Hörstadius S. Über die Determination im Verlaufe der Eiachse bei Seeigeln. Pubbl Stn Zool Napoli. 1935;14:251–479. doi: 10.1007/BF02569028. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. A complete second gut induced by transplanted micromeres in the sea urchin embryo. Science. 1993;259:1134–1138. doi: 10.1126/science.8438164. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. Micromeres are required for normal vegetal plate specification in sea urchin embryos. Development. 1995;121:3215–3222. doi: 10.1242/dev.121.10.3215. [DOI] [PubMed] [Google Scholar]
- Ishizuka Y, Minokawa T, Amemiya S. Micromere descendants at the blastula stage are involved in normal archenteron formation in sea urchin embryos. Dev Genes Evol. 2001;211:83–88. doi: 10.1007/s004270000120. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci U S A. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 2007;318:794–797. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- Wei Z, Angerer LM, Gagnon ML, Angerer RC. Characterization of the SpHE promoter that is spatially regulated along the animal-vegetal axis of the sea urchin embryo. Dev Biol. 1995;171:195–211. doi: 10.1006/dbio.1995.1271. [DOI] [PubMed] [Google Scholar]
- Howard EW, Newman LA, Oleksyn DW, Angerer RC, Angerer LM. SpKrl: a direct target of beta-catenin regulation required for endoderm differentiation in sea urchin embryos. Development. 2001;128:365–375. doi: 10.1242/dev.128.3.365. [DOI] [PubMed] [Google Scholar]
- Livi CB, Davidson EH. Expression and function of blimp1/krox, an alternatively transcribed regulatory gene of the sea urchin endomesoderm network. Dev Biol. 2006;293:513–525. doi: 10.1016/j.ydbio.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, et al. Nuclear Beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev Biol. 2002;246:132–147. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by FoxA, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- Gross JM, McClay DR. The role of Brachyury(T) during gastrulation movements in the sea urchin Lytechinus variegatus. Dev Biol. 2001;239:132–147. doi: 10.1006/dbio.2001.0426. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Lapraz F, Rottinger E, Duboc V, Range R, Duloquin L, et al. RTK and TGF-β signaling pathways genes in the sea urchin genome. Dev Biol. 2006;300:132–152. doi: 10.1016/j.ydbio.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Rottinger E, Besnardeau L, Lepage T. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell. 2004;6:397–410. doi: 10.1016/s1534-5807(04)00056-5. [DOI] [PubMed] [Google Scholar]
- Range R, Lapraz F, Quirin M, Marro S, Besnardeau L, et al. Cis-regulatory analysis of Nodal and maternal control of dorsal-ventral axis formation by Univin, a TGF-ß related to Vg1. Development. 2007;134:3649–3664. doi: 10.1242/dev.007799. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Duboc V, Eric R, Lapraz F, Lydia B, Thierry L. Left-right asymmetry in the sea urchin embryo is regulated by Nodal signaling on the right side. Dev Cell. 2005;9:147–158. doi: 10.1016/j.devcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Sweet HC, Gehring MC, Ettensohn CA. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. Cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Minokawa T, Amemiya S. Timing of the potential of micromere-descendants in echinoid embryos to induce endoderm differentiation of mesomere-descendants. Develop Growth Differ. 1999;41:535–547. doi: 10.1046/j.1440-169x.1999.00453.x. [DOI] [PubMed] [Google Scholar]
- Amore G, Yavrouian RG, Peterson KJ, Ransick A, McClay DR, et al. Spdeadringer, a sea urchin embryo gene required separately in skeletogenic and oral ectoderm gene regulatory networks. Dev Biol. 2003;261:55–81. doi: 10.1016/s0012-1606(03)00278-1. [DOI] [PubMed] [Google Scholar]
- Kenny AP, Kozlowski DJ, Oleksyn DW, Angerer LM, Angerer RC. SpSoxB1, a maternally encoded transcription factor asymmetrically distributed among early sea urchin blastomeres. Development. 1999;126:5473–5483. doi: 10.1242/dev.126.23.5473. [DOI] [PubMed] [Google Scholar]
- Kenny AP, Oleksyn DW, Newman LA, Angerer RC, Angerer LM. Tight regulation of SpSoxB factors is required for patterning and morphogenesis in sea urchin embryos. Dev Biol. 2003;261:412–425. doi: 10.1016/s0012-1606(03)00331-2. [DOI] [PubMed] [Google Scholar]
- Hörstadius S. Experimental embryology of echinoderms. Oxford (United Kingdom): Clarendon Press; 1973. 192 [Google Scholar]
- Ransick A, Davidson EH. Late specification of veg1 lineages to endodermal fate in the sea urchin embryo. Dev Biol. 1998;195:38–48. doi: 10.1006/dbio.1997.8814. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Harger P, Mitchell A, Lemaire P. Activin signalling and response to a morphogen gradient. Nature. 1994;371:487–492. doi: 10.1038/371487a0. [DOI] [PubMed] [Google Scholar]
- D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Yasunaga M, Tada S, Torikai-Nishikawa S, Nakano Y, Okada M, et al. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- Piepenburg O, Grimmer D, Williams PH, Smith JC. Activin redux: specification of mesodermal pattern in Xenopus by graded concentrations of endogenous ActivinB. Development. 2004;131:4977–4986. doi: 10.1242/dev.01323. [DOI] [PubMed] [Google Scholar]
- Ramis JM, Collart C, Smith JC. Xnrs and Activin regulate distinct genes during Xenopus development: Activin regulates cell division. PLOS One. 2007;2:e213. doi: 10.1371/journal.pone.0000213. doi: 10.1371/journal.pone.0000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann CE, Hemmati-Brivanlou A, Thomsen GH, Fields A, Woolf TM, et al. Expression of Activin mRNA during early development in Xenopus laevis . Dev Biol. 1993;157:474–483. doi: 10.1006/dbio.1993.1150. [DOI] [PubMed] [Google Scholar]
- Livingston BT, Wilt FH. Range and stability of cell fate determination in isolated sea urchin blastomeres. Development. 1990;108:403–410. doi: 10.1242/dev.108.3.403. [DOI] [PubMed] [Google Scholar]
- Rui Y, Xu Z, Xiong B, Cao Y, Lin S, et al. A β-Catenin-independent dorsalization pathway activated by Axin/JNK signaling and antagonized by Aida. Dev Cell. 2007;13:268–282. doi: 10.1016/j.devcel.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Minokawa T, Wikramanayake AH, Davidson EH. Cis-regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Dev Biol. 2005;288:545–558. doi: 10.1016/j.ydbio.2005.09.047. [DOI] [PubMed] [Google Scholar]