Abstract
It is commonly thought that disruption of the circadian clock increases the cancer incidence in humans and mice. However, it was found that disruption of the clock by the Cryptochrome (Cry) mutation in mice did not increase cancer rate in the mutant mice even after exposing the animals to ionizing radiation. Therefore, in this study we tested the effect of the Cry mutation on carcinogenesis in a mouse strain prone to cancer because of a p53 mutation, with the expectation that clock disruption in this sensitized background would further increase cancer risk. Paradoxically, we find that the Cry mutation protects p53 mutant mice from the early onset of cancer and extends their median lifespan ≈50%, in part by sensitizing p53 mutant cells to apoptosis in response to genotoxic stress. These results suggest alternative therapeutic approaches in management of cancers associated with a p53 mutation.
Keywords: apoptosis, circadian clock, DNA repair
Circadian rhythm is the cyclical change in the physiology and behavior of organisms with a periodicity of ≈24 h (1). This rhythm is generated by a molecular clock that is present in virtually all cells in humans and mice. The peripheral clocks throughout the organism are coordinated by a master clock located in the suprachiasmatic nuclei (SCN) in the hypothalamus. Clock disruption by environmental or genetic factors has been implicated in a number of pathologic conditions (2–9). Of interest, several epidemiologic studies and, more recently, some studies with animal model systems have suggested that circadian clock disruption predisposes humans and mice to cancer (5, 6).
The significant progress that has been made recently in understanding the circadian clock at the molecular level (10, 11) has provided the conceptual and technical tools for investigating the clock disruption-cancer connection at a mechanistic level. The mammalian circadian clock is generated by a transcription-translation feedback loop (TTFL): The transcriptional activators Clock and BMal1 proteins make a heterodimer that binds to the promoter of the transcriptional repressor genes Cryptochrome 1 and 2 (Cry1 and Cry2) and Period 1 and 2 (Per 1 and Per2) and induces their transcription. The Cry and Per proteins make heterodimers, which, after a time lag, inhibit the Clock-BMal1 complex and hence their own transcription (10–12). The time delay between the synthesis of the Per and Cry and their action as repressors is a key factor in generating an oscillatory pattern of expression of these proteins and of the clock controlled genes that produce the circadian phenotype (12). Although this basic circuitry is consolidated by posttranslational modifications and by interfacing with other transcriptional circuits, Clock, BMal1, Crys and Pers are considered the core clock genes. Hence studies aimed at investigating the clock disruption-cancer connection have either targeted these genes or used animals in which the master circadian clock in the SCN was surgically destroyed.
It was found that in SCN-lesioned arrhythmic mice, subcutaneous (s.c.) implants of 2 different types of cancers grew at a faster rate than in the controls with normal circadian rhythm (13), leading to the conclusion that circadian disruption at the organism level facilitated tumor growth. This conclusion was reinforced by a study with Per2 mutant mice. It was reported that these mice developed spontaneous and ionizing radiation (IR)–induced lymphomas at a much higher rate than the wild-type controls (14). These studies, combined with the epidemiological data on humans, led to the generalization that circadian clock disruption by any means predisposes animals to cancer. However, subsequent work with mice with mutations in either the negative arm (Cry) or the positive arm (Clock) of the TTFL did not support this generalization. It was found that Cry1−/−Cry2−/− (15) and clock/clock (16) mutant mice were indistinguishable from wild-type mice with respect to the incidence of spontaneous and IR-induced tumors. Thus, it was concluded that not the circadian clock disruption in itself but the manner in which the clock is disrupted is more germane to a potential increase in cancer susceptibility. However, it was also conceivable that the studies with the Cry and Clock mutants were inconclusive with respect to the long-term effects of circadian disruption, in particular when clock disruption occurs in a cancer sensitized genetic background such as occurs in animals with a mutation in either an oncogene or a tumor suppressor. To address this particular issue we decided to combine the Cry mutation with the p53 mutation.
The tumor suppressor p53 is mutated in nearly half of human cancers (17). In mouse models for human cancers, often p53 mutations are combined with mutations in other genes that are suspected to play a role in cancer development (18). Commonly, such combinations increase the incidence of cancer that is already quite high in p53 mutant mice (19–22). Paradoxically, we found that the Cry mutation delays cancer onset in p53 mutant mice and increases the median lifespan of these mice ≈1.5-fold. This finding raises the possibility of cancer management by interfering with cryptochrome function at the level of the whole organism or selectively in cancer tissue.
Results
Generation of p53−/−Cry1−/−Cry2−/− Mice.
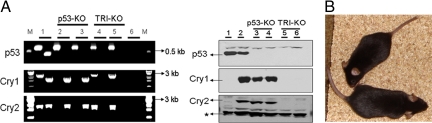
We crossed p53+/− mice in C57BL/6J background (23) with Cry1+/−Cry2+/− mice in the same background (15), and by interbreeding the progeny of this cross we obtained the p53−/−Cry1−/−Cry2−/− mice and mice with other combinations of p53 and Cry mutations. As is well known for p53−/− mice (24), there was a strong bias for males among the triple knockout progeny (male:female = 19:2). Fig. 1A shows the genotyping of the mutants by genomic PCR and immunoblotting for p53, Cry1, and Cry2 proteins. Fig. 1B shows that a comparison of 8-week old p53−/− and p53−/−Cry1−/−Cry2−/−mice reveals no obvious physical differences between the 2 mice except the triple knockouts weigh ≈20% less than the p53−/− mice, an observation that has been made previously in comparing Cry1−/−Cry2−/− with their wild-type littermates (25).
Fig. 1.
Genotyping and physical appearance of mice used in this study. (A) Isolated skin fibroblasts from 2 age-matched mice of p53−/− (p53-KO) and p53−/−Cry1−/−Cry2−/− (TRI-KO) genotypes were used for analysis. (Left) Genotyping by genomic PCR. For each gene, an anchor and a knockout or wild-type allele-specific primers were used. Genomic DNA from the tail of a triple heterozygous mouse was used as a positive control for each set of primer pairs. Lane 1, DNA from a triple heterozygous mouse; lanes 2 and 3, DNA from 2 p53-KO fibroblasts; lanes 4 and 5, DNA from 2 TRI-KO fibroblasts; lane 6, no genomic DNA. M, molecular size markers. (Right) Immunoblot of fibroblast lysates. 50 μg of protein from lysates of cultured fibroblasts were used for immunoblotting. Lanes 1 and 2, lysates from immortalized Cry1−/−Cry2−/− and wild-type (WT) fibroblasts, respectively (15); lanes 2 and 3, lysates from fibroblasts shown in Left lanes 2 and 3; lanes 5 and 6, lysates from fibroblasts shown in Left lanes 4 and 5. A protein band cross-reacting with Cry2 antibody (indicated by *) is shown as a loading control. (B) Physical appearance of mutant mice. TRI-KO (top) and p53-KO (bottom) at 8 weeks of age appear anatomically normal but the TRI-KO mutants are smaller and weigh ≈20% less than the p53-KO mice.
Cancer Incidence in Cryptochrome-deficient p53−/− Mice.
It is well-established that p53−/− mice develop cancers, mostly lymphomas and lymphosarcomas, with a median age of ≈20 weeks in C57BL/6J background (18, 24). Combination of p53−/− with a mutation in a tumor suppressor such as Rb (20) or with an up-regulated oncogene, such as ras or myc (19, 22), increases the incidence of cancer. Surprisingly, even combinations of p53−/− with mutations that were expected to reduce cancer incidence did not afford protection to p53−/− mice (26).
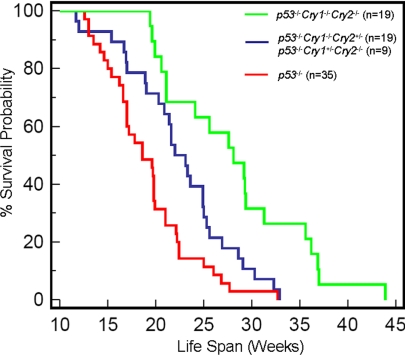
Hence it appears that p53 null mutations have a rather penetrant phenotype that may be amplified under certain circumstances but that rarely, if ever, can be overcome by other mutations. Against this background then, the effect of Cry null mutations on p53−/− mice was surprising. As seen in Fig. 2, the age adjusted incidence of cancer in p53−/−Cry1−/−Cry2−/− mice was significantly lower than that of p53−/− mice and the median lifespan of the triple knockout mice (28.1 weeks) was nearly 1.5-fold that of the p53−/− mice (18.6 weeks). Even mutating 3 out of the 4 Cry genes provided some protection as apparent from a lifespan of 23.1 weeks for p53−/−Cry1+/−Cry2−/− and p53−/−Cry−/−Cry2+/− mice (Fig. 2). It is known that a single Cry gene provides partial molecular and behavioral rhythmicity in mice (27–29). Thus, it appears that reducing the level of or eliminating cryptochrome imparts protection from cancer death to p53−/− mice in a manner proportional to the degree of cryptochrome reduction.
Fig. 2.
Mutations of Cryptochromes in p53−/− mice reduces age adjusted tumor incidence. Kaplan–Meier survival analysis (log-rank test) of the time of death with evidence of tumors showed significant differences between p53−/− and p53−/−Cry1−/−Cry2+/− or p53−/−Cry1+/−Cry2−/− (P = 0.0148), between p53−/− and p53−/−Cry1−/−Cry2−/− (P < 0.0001), and between p53−/−Cry1−/−Cry2+/− or p53−/−Cry1+/−Cry2−/− and p53−/−Cry1−/−Cry2−/− (P = 0.0043). There was no significant survival difference between p53−/−Cry1−/−Cry2+/− (n = 19) and p53−/−Cry1+/−Cry2−/− (n = 9) (P = 0.0638) and hence these groups were combined in our analysis.
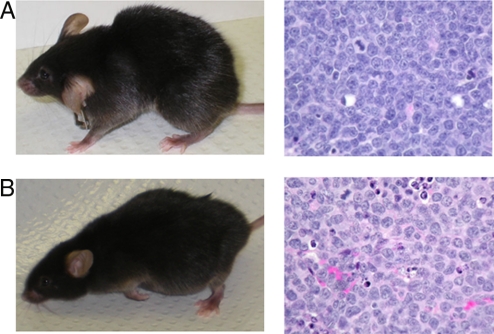
However, the triple knockout mice do eventually succumb to death from cancer. Histopathological examination reveals that these mice have a delayed development of the typical lymphoblastic lymphomas seen in p53−/− mice. Histopathological types of some of the tumors (such as non-lymphoblastic lymphoma or histiocytic sarcoma) are those associated with late developing lymphomas in p53−/− mice or commonly observed in aged mice (Fig. 3). This raises the question as to why the triple knockout mice have a lower age-adjusted incidence of cancer and longer lifespan than the p53−/− mice. This question is addressed below.
Fig. 3.
Histopathological analyses of mice with tumors. Both p53-KO and TRI-KO mice develop thymic lymphoma, often coupled with peripheral lymphadenopathy. In addition, TRI-KO mice also present abdominal distension due to massive lymphoma and other sarcomas in the liver and spleen typically seen in older p53-KO mice. (A) (Left) A representative 28-week old p53-KO mouse with thymic lymphoblastic lymphoma. (Right) A hematoxylin and eosin–stained section of thymic lymphoma from this mouse. (B) (Left) A representative TRI-KO mouse at 36 weeks of age exhibiting an abdominal mass in addition to the thymic lymphoma. (Right) A hematoxylin and eosin–stained section of thymic lymphoma from this mouse.
Molecular Analysis of Carcinogenesis in p53−/− Mice Without Cryptochrome.
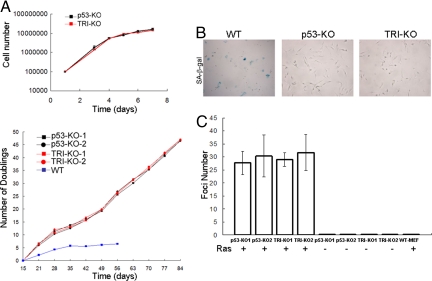
The reduced rate of cancer incidence in, and the longer lifespan of, the triple knockout mice relative to the p53−/− mice by the Cry mutation could be due to either prevention (or delay) of malignant transformation of p53−/− cells or activation of some signaling pathways that eliminate transformed cells and that have been abrogated in p53−/− cells. To differentiate between these possibilities, we isolated skin fibroblasts from p53−/− and p53−/−Cry1−/−Cry2−/− mice and tested them for a variety of cell growth and proliferation endpoints. p53 is a multifunctional protein that prevents cancer by promoting cell cycle arrest, senescence, or apoptosis in response to genotoxic stress and to proliferative signals such as those given by oncogenes (17, 18). Hence, we tested the cell lines from the triple knockout mutant mice to find out which of these response reactions lost by p53 mutation may have been restored by the Cry mutation. First, we analyzed the growth properties of skin fibroblasts isolated from p53−/− and the triple knockout mutants. Fig. 4A shows that the fibroblasts of the 2 genotypes grow at the same rate (Upper), and, when tested for multiple passages, they appear to be already immortalized, in contrast to the fibroblasts from wild-type mice, which slow down dividing after ≈5 doublings (Lower). In parallel with these findings the p53−/− and the triple knockout fibroblasts show no evidence of senescence as measured by the senescence-associated β galactosidase assay, while normal fibroblasts senesce under these conditions (Fig. 4B). Thus, we conclude that elimination of cryptochrome does not affect the proliferative phenotype of p53−/− cells nor does it restore the senescence phenotype that has been lost in these cells.
Fig. 4.
Analyses of p53-KO and TRI-KO fibroblasts for growth, immortalization, senescence, and malignant transformation properties. (A) (Upper) Growth kinetics of the fibroblasts. Cells were seeded into 6-well plates and counted after incubation for the indicated times. (Lower) Long-term cumulative population doubling assay for fibroblasts were determined using cells passaged every 3–4 days. Cells were seeded into 6-well plates and counted after 48 h. Population doublings were calculated at different passages and plotted. Fibroblasts from a wild-type mouse were maintained for at least 8 passages without any overt immortalization. (B) Senescence-associated β galactosidase staining (blue) of fibroblasts at late passages (passage 25 for p53-KO and TRI-KO). Wild-type skin fibroblasts (passage 8) from an age-matched mouse were used as a positive control. (C) Ras-induced oncogenic transformation. Fibroblasts from 2 p53-KO mice (p53-KO1 and p53-KO2) and 2 triple mutant mice (TRI-KO1 and TRI-KO2) were transfected with Ras pT24+pEGFP.N1 (+) or pEGFP.N1 (-), and colonies were counted after 19 days. Colony numbers were adjusted to transfection efficiencies, which were quite similar for p53-KO and TRI-KO fibroblasts. WT-MEF, wild-type mouse embryonic fibroblasts were used as a negative control. Bars indicate standard errors of the mean.
Then, we tested the primary skin fibroblasts for foci formation induced by Ras to probe for oncogenic transformability of the fibroblasts (30). The data shown in Fig. 4C reveal that while normal fibroblasts fail to produce Ras-induced foci, 2 independent isolates of both p53−/− and p53−/−Cry1−/−Cry2−/− fibroblasts produce foci at the same frequency upon transfection with the Ras oncogene. We conclude that removal of cryptochrome in a p53−/− background does not prevent immortalization of the cell nor does it prevent its oncogenic transformation. Therefore, the reduced incidence of overt cancers and the increased lifespan of the triple knockout mice may have been caused by an abnormal response to inevitable genotoxic stress, to which oncogenically transformed cells are subjected. In the following series of experiments we tested for this response.
Genotoxic Responses of p53−/− and Cryptochromeless p53−/− Skin Fibroblasts.
To analyze the contribution of cryptochrome to the response of p53−/− cells to genotoxic stress, we irradiated p53−/− and p53−/−Cry1−/−Cry2−/− mouse skin fibroblasts with UV and measured clonogenic survival, DNA repair, DNA damage checkpoint response, and apoptosis. We found that the elimination of cryptochrome increases the sensitivity of p53−/− cells to killing by UV (Fig. 5). To determine whether the increased sensitivity was due to reduced repair capacity, defective DNA damage checkpoint response, or enhanced apoptosis, we analyzed these responses to DNA damage in p53−/− and triple knockout cell lines. We found that elimination of cryptochrome does not affect the nucleotide excision repair capacity of the fibroblasts as measured by cyclobutane thymine dimer removal in vivo (Fig. 6A) and (6–4) photoproduct removal in vivo (data not shown) and with cell free extracts (Fig. 6B). Similarly, although the Cry deletion led to an increase in the level of the antimitotic kinase Wee1, as reported previously (15, 32, 33) (Fig. 7A), it had no effect on the G2/M checkpoint response, as reported previously (15), nor on normal functioning of the ATR → Chk1 and ATM → Chk2 signaling pathways (Fig. 7B). In contrast, we found that p53−/−Cry1−/−Cry2−/− cells are more susceptible to UV-induced apoptosis than p53−/− cells as measured by proteolytic cleavage of caspase 3 and Poly (ADP) ribose polymerase (PARP) (Fig. 7 C and D), which likely explains the increased sensitivity to UV killing of the triple knockout fibroblasts relative to the p53−/− fibroblasts. The potential significance of these findings to the increased lifespan of the p53−/−Cry1−/−Cry2−/− mice relative to p53−/− mice is discussed below.
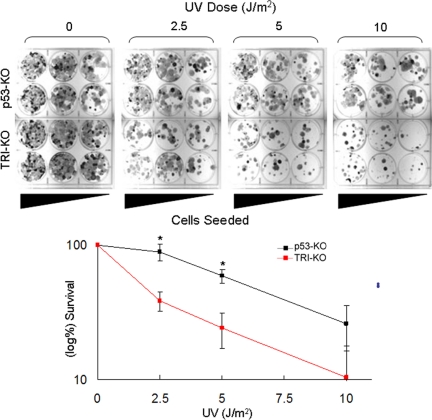
Fig. 5.
Clonogenic survival assay for primary skin fibroblasts from p53-KO and TRI-KO mice. (Upper) Into 6-well plates, 105, 5 × 104, and 2 × 104 cells were seeded in duplicates, irradiated with UV, and then incubated for 9–10 days until colonies were readily visible. Colonies were then stained with 5% methylene blue. (Lower) UV survival curves obtained from 3 experiments including the 1 shown in Upper. Error bars indicate standard errors of the mean and the asterisks indicate statistically significant (P < 0.01) difference in clonogenic survival.
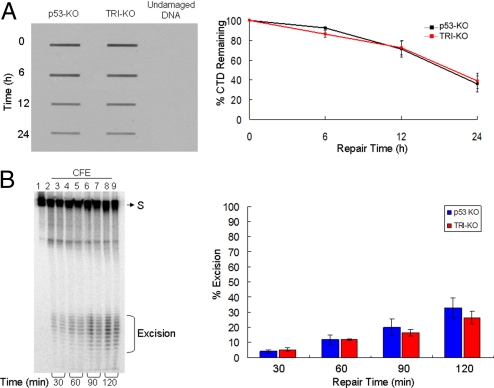
Fig. 6.
In vivo and in vitro excision repair capacity of p53-KO and TRI-KO fibroblasts. (A) In vivo repair assay. Fibroblasts of the indicated genotypes were exposed to a 10 J/m2 UV dose and the level of repair was quantified by slot-blot with anti-CTD monoclonal antibody. (Left) A representative assay. (Right) Quantitative analysis of data from 3 independent experiments including the 1 shown in Left. The repair kinetics of CTD and of (6–4) photoproducts (data not shown) of p53−/− fibroblasts are in agreement with a previous report (31). (B) (Left) Removal of (6–4) photoproducts in vitro with cell free extracts (CFE). Extracts from p53-KO and TRI-KO cell lines were incubated with a 136-bp substrate (S) containing a (6–4) T-T photoproduct, for the indicated times. Reaction products were then analyzed on a 10% sequencing gel. The excision products in the 24–32 nt range are bracketed. Lane 1, substrate without treatment; lanes 2, 4, 6, and 8, substrates treated with p53-KO fibroblast CFE for 30, 60, 90, and 120 min; lanes 3, 5, 7, and 9, substrates treated with TRI-KO CFE for 30, 60, 90, and 120 min. (Right) Quantitative analysis of the excision from 3 experiments including the 1 shown in Left. Error bars indicate standard errors of the mean. There was no statistically significant difference (P > 0.05 by a 2-sample t test) in the excision activities of the 2 extracts.
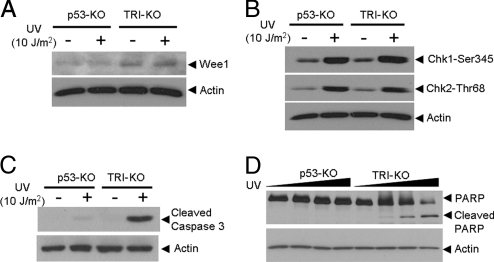
Fig. 7.
Analyses of p53-KO and TRI-KO fibroblasts for cell cycle regulation, DNA damage checkpoints, and apoptosis, after a genotoxic treatment. The indicated cell lines were irradiated and probed for various damage response endpoints. (A) Wee-1 expression is up-regulated in triple knockout fibroblasts, which is in agreement with previously published data (15), and this level is not affected by UV. (B) Both the ATR → Chk1 and ATM → Chk2 checkpoint pathways appear to be functioning normally in the absence of cryptochrome, as evidence by phosphorylation of signal transducing kinases after UV treatment. Dose-response (2 h time point after 0, 2.5, 5, and 10 J/m2 of UV treatment) and kinetic experiments (1, 2, and 4 h after 5 J/m2 of UV treatment) did not reveal any difference in Chk1 and Chk2 phosphorylation in fibroblasts of the 2 genotypes (data not shown). (C, D) Assay for UV-induced apoptosis. The p53-KO and TRI-KO fibroblasts were irradiated with UV and tested for apoptosis by caspase 3 cleavage (24 h) and PARP cleavage, 24 h after UV dose treatments at 0, 10, 20, and 50 J/m2. Cell lysates were also probed for actin as a loading control.
Discussion
Circadian Cycle and Cell Cycle.
The circadian clock and the cell cycle are 2 global regulatory systems at cellular and organismic levels and therefore are expected to interface. Indeed, the systems are known to interface at a number of critical points. BMal1-Clock positively regulates the Wee1 antimitotic kinase (15, 32, 33), and BMal1-Clock or BMal1-NPas2 represses c-Myc transcription (14). The transcription of c-Myc was reported to be highly elevated in Per2 mutant mice (because Per2 positively regulates BMal1 transcription and in the absence of Per2 the BMal1, a repressor of c-Myc, is down) and the high incidence of IR-induced lymphoma in these mice was ascribed, at least in part, to the elevated c-Myc (14). In addition to the circadian cycle-cell cycle connection at the transcriptional level, the 2 systems interface at the protein–protein interaction and signal modulation level as well. First, the Timeless protein, which is required for a robust circadian rhythm in mice (34), directly interacts with the cell cycle checkpoint proteins ATR and Chk1 such that down-regulation of Timeless interferes with the ATR → Chk1 branch of the DNA damage checkpoint response (35). Second, Per1 has been reported to interact with both the ATM and Chk2 proteins and to directly participate in the ATM → Chk2 DNA damage signaling pathway (36). Moreover, it has been reported that Per1 overexpression induces cell death in several cancer cell lines, presumably by activating the ATM → Chk2 signaling axis and arresting cell proliferation and promoting apoptosis (36). Clearly, these are interesting findings that need further expansion before they can be considered as potential circadian clock-checkpoint connections that might be exploited in cancer management.
Circadian Cycle and Cancer.
Despite the epidemiological findings suggesting that disruption of the circadian clock predisposes humans to cancer (5, 37, 38), currently available data from mouse model systems reveal a rather conflicting picture. Disruption of the clock by the Per2 mutation is reported to predispose mice to cancers (14), but disruption of the clock by the Cry mutation (which, like the Per2 mutation, affects the negative arm of the clock TTFL) does not (15), nor does the mutation of Clock, which affects the positive arm of the circadian TTFL (16). BMal1 mutants exhibit an early aging phenotype and die at a relatively young age (16, 39) and, hence, are not suitable for studying carcinogenesis. Taking into account the data from Per2, Cry, and Clock mutants, the only conclusion that can be made at present is that clock disruption per se does not predispose mice to cancers and that these core clock genes must participate in functions that are unique to each and are separable from the clock functions of these genes to give rise to such different phenotypes.
Cancer Management by Clock Disruption.
The results presented in this article taken at face value would suggest that down-regulating Cry or interfering with Cry function in cancers associated with p53 null mutations may slow the progression of cancer and, in combination with other approaches to cancer treatment, it may increase the success rate of remission. At present we do not know how the Cry mutation renders oncogenically transformed p53−/− cells more susceptible to apoptosis and delays the appearance of overt tumors. One possibility supported by our data is that the deletion of Cry makes the oncogenically transformed cells more sensitive to apoptotic signals. Interestingly, even though Wee1 (which is elevated in Cry mutants) is well known for its effect on mitosis as the enzyme that phosphorylates and hence inhibits mitotic kinase Cdc2, it has also been found that it specifically interacts with the SH2 domain of the Crk adaptor protein and, in doing so, activates the apoptosis signaling cascade both in Xenopus egg extract (40) and in the human U2OS osteosarcoma line (41). In addition to this potential role of Wee1 in apoptosis, it is conceivable that the Cry mutation may delay tumorigenesis by affecting non–cellautonomous factors, such as altered immune surveillance of tumors. It is also possible that the Cry mutation reduces tumorigenicity in p53−/− mice by reducing the level of reactive oxygen species that are known to be elevated in BMal1−/− mice (39) and could conceivably be reduced in Cry mutants. Clearly, further work is needed to provide mechanistic explanations for the reduction of cancer risk in p53−/− mice by the Cry mutation.
Materials and Methods
Generation of p53−/−Cry1−/−Cry2−/− Mice.
p53+/− mice in C57BL/6J background (23) and Cry1+/−Cry2+/− mice with the same background (15) were used to obtain p53−/− mice, mice with p53−/− and single Cry, and the triple knockout mice. Most of triple knockouts were generated from crossing p53−/− Cry1+/−Cry2+/− males and p53+/−Cry1+/−Cry2+/− females. Genotyping was done before the age of 19 days. Genomic DNA was isolated from the tail for PCR. For each gene, an anchor and a primer specific for knockout or wild-type sequences were used. PCR products were analyzed on a 1% (wt/vol) agarose gel. The genotypes of immortalized skin fibroblasts isolated from some of the mutants were confirmed by immunoblotting.
Isolation of Fibroblasts and Growth Curves.
We used skin fibroblasts from 2 age matched p53−/− and triple knockout mice for molecular analyses. Skin patches from the back of mice at the age of 3 months were taken under sterile conditions, cut into small pieces, placed into 4 or 6 well-plates, and incubated 1 week to let the fibroblasts migrate into the wells. Fibroblasts were passaged at least 3 times before using them for any experiment. Wild-type mouse embryonic fibroblasts were kindly provided by Dr. Norman Sharpless (University of North Carolina). Primary wild-type skin fibroblasts were isolated as described above, and passaged 8 times without appearance of spontaneously immortalized clones. For the population doubling (PD) assay, 50 × 104 cells were plated into 6-well plates, and counted after 48 h, using a hemocytometer. PD was determined by the formula: PD = Log (Nf/Ni)/Log2, where Nf = the number of cells counted and Ni = the number of cells seeded. Senescence associated β-galactosidase (SABG) activity was detected using a standard protocol (42).
Assay for Ras-induced Oncogenic Transformation.
Cells were seeded into 10 cm plates, and transfected with 250 ng of pEGFP-N1 (Clontech), and/or 5 μg of Ras pT24 vector (30) using FuGENE 6 reagent (Roche). The next day, cells were split into 3 plates, and incubated for 18 days. The plates were stained with Giemsa and colonies of oncogenically transformed cells were counted visually and the fraction of transformed cells was normalized for transfection efficiencies calculated by counting GFP positive cells in each plate. The results shown are from 3 independent transfections.
Histopathological Analyses.
Animals were handled according to the guidelines of the National Institutes of Health and the University of North Carolina. Animals with visible tumors or exhibiting severe morbidity were euthanized with carbon dioxide and fresh tissues were taken for pathological examination. Parts of some tissues were fixed in 10% buffered formalin, and submitted to the University of North Carolina School of Medicine Pathology Department for analysis.
Treatment of Cells with UV Radiation for Survival Assay.
Fibroblasts of indicated genotypes were seeded into 6-well plates at 105, 5 × 104, and 2 × 104 cells per well in duplicates, and exposed to 10 J/m2 of UV (254 nm) radiation from a germicidal lamp at a fluence rate of 0.5 Jm−2s−1. After UV treatment, the cells were washed with PBS and incubated in high-glucose DMEM for 9 to 10 days until colonies were readily visible. Cells were fixed for 20 min in 3:1 methanol/acetic acid, rinsed with water, and stained with 5% methylene blue. Colonies containing > 50 cells were scored.
Measurement of DNA Repair Activity In Vivo.
Exponentially growing cells were treated with UV (254 nm) radiation at a dose of 10 J/m2. Cells were harvested at 0, 6, 12, and 24 h after UV radiation and stored at −20 °C until processing. Genomic DNA was isolated using the QIAamp DNA Mini kit (Qiagen). Cyclobutane thymine dimers (CTDs) were detected by slot blot using anti-CTD mouse monoclonal antibody (Kamiya Biomedical) diluted in PBS (1:2,000) containing 0.1% Tween 20 for 16 h at 4 °C, which was followed by incubation with an anti-mouse antibody-HRP conjugate (43). CTD signals were detected using an ECL reagent (GE Healthcare). Quantification was performed with ImageQuant 5.2 software (Molecular Dynamics).
Measurement of DNA Repair Activity In Vitro.
The excision repair assay (44) was carried out with a 136-bp DNA duplex containing a (6–4) photoproduct, as described elsewhere (45), using cell-free extracts prepared from p53−/− and p53−/−Cry1−/−Cry2−/− primary fibroblasts that were supplemented with recombinant RPA. The excision levels were quantified using ImageQuant 5.2 software (Molecular Dynamics).
DNA Damage Checkpoint and Apoptosis Assays.
For apoptosis analysis using caspase 3 cleavage, cells were treated with 10 J/m2 UV dose, and incubated for 24 h. For apoptosis analysis using PARP cleavage, cells were treated with 10, 20, 50 J/m2 UV doses and incubated for 24 h. For checkpoint response, cells were treated with 10 J/m2 UV dose and incubated for 2 h. For estimating Wee1 levels, phosphorylation status of Chk1 and Chk2, and proteolytic cleavage of caspase 3 and PARP, 50 μg of protein from total cell lysates were used for immunoblotting with the appropriate antibodies. Rabbit polyclonal anti-Wee1 (H-300), goat polyclonal anti-actin, rabbit polyclonal anti-p53, and anti-IgG goat-HRP conjugates were obtained from Santa Cruz Biotechnology. Rabbit polyclonal anti-phospho-ser345-Chk1, anti-phospho-thr68-Chk2, cleaved caspase 3, and PARP antibodies were from Cell Signaling Technology, anti-IgG rabbit-HRP conjugates were from GE HealthCare, and rabbit polyclonal anti-Cry2 was from Alpha Diagnostic International. Rat monoclonal anti-Cry1 (IgM) antibody was produced in our laboratory (15).
Acknowledgments.
We thank Norman Sharpless and Christine Petre for their critical advice on analyses of fibroblasts and for supplying the Ras pT24 vector and wild-type MEFs, Randy J. Thresher and Stephen Knight for their help with skin fibroblast isolation, and Virginia L. Godfrey for histopathological analysis of tumor samples. We thank Drs. David Mu (Pennsylvania State University), Mehmet Öztürk (Bilkent University), and Russel Van Gelder (University of Washington) for useful comments. This work was supported by National Institutes of Health grant GM31082.
Footnotes
The authors declare no conflict of interest.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz WJ. A clinician's primer on the circadian clock: Its localization, function, and resetting. Adv Intern Med. 1993;38:81–106. [PubMed] [Google Scholar]
- 3.Sancar A. Cryptochrome: The second photoactive pigment in the eye and its role in circadian photoreception. Annu Rev Biochem. 2000;69:31–67. doi: 10.1146/annurev.biochem.69.1.31. [DOI] [PubMed] [Google Scholar]
- 4.Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 5.Stevens RG. Circadian disruption and breast cancer: From melatonin to clock genes. Epidemiology. 2005;16:254–258. doi: 10.1097/01.ede.0000152525.21924.54. [DOI] [PubMed] [Google Scholar]
- 6.Fu L, Lee CC. The circadian clock: Pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi M, Wood PA, Hrushesky WJ. Circadian chemotherapy for gynecological and genitourinary cancers. Chronobiol Int. 2002;19:237–251. doi: 10.1081/cbi-120002600. [DOI] [PubMed] [Google Scholar]
- 8.Kondratov RV, Antoch MP. Circadian proteins in the regulation of cell cycle and genotoxic stress responses. Trends Cell Biol. 2007;17:311–317. doi: 10.1016/j.tcb.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 10.Hughes M, et al. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb Symp Quant Biol. 2007;72:381–386. doi: 10.1101/sqb.2007.72.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueda HR. Systems biology of mammalian circadian clocks. Cold Spring Harb Symp Quant Biol. 2007;72:365–380. doi: 10.1101/sqb.2007.72.047. [DOI] [PubMed] [Google Scholar]
- 12.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipski E, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–697. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 14.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 15.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 16.Antoch MP, et al. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–1204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attardi LD, Jacks T. The role of p53 in tumour suppression: lessons from mouse models. Cell Mol Life Sci. 1999;55:48–63. doi: 10.1007/s000180050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attardi LD, Donehower LA. Probing p53 biological functions through the use of genetically engineered mouse models. Mutat Res. 2005;576:4–21. doi: 10.1016/j.mrfmmm.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 19.Blyth K, et al. Synergy between a human c-myc transgene and p53 null genotype in murine thymic lymphomas: Contrasting effects of homozygous and heterozygous p53 loss. Oncogene. 1995;10:1717–1723. [PubMed] [Google Scholar]
- 20.Williams BO, et al. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat Genet. 1994;7:480–484. doi: 10.1038/ng0894-480. [DOI] [PubMed] [Google Scholar]
- 21.Donehower LA, et al. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 1995;9:882–895. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- 22.Hundley JE, et al. Increased tumor proliferation and genomic instability without decreased apoptosis in MMTV-ras mice deficient in p53. Mol Cell Biol. 1997;17:723–731. doi: 10.1128/mcb.17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simin K, et al. Deciphering cancer complexities in genetically engineered mice. Cold Spring Harb Symp Quant Biol. 2005;70:283–290. doi: 10.1101/sqb.2005.70.038. [DOI] [PubMed] [Google Scholar]
- 24.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 25.Tamanini F, Chaves I, Bajek MI, van der Horst GT. Structure function analysis of mammalian cryptochromes. Cold Spring Harb Symp Quant Biol. 2007;72:133–139. doi: 10.1101/sqb.2007.72.066. [DOI] [PubMed] [Google Scholar]
- 26.Nacht M, Jacks T. V(D)J recombination is not required for the development of lymphoma in p53-deficient mice. Cell Growth Differ. 1998;9:131–138. [PubMed] [Google Scholar]
- 27.Thresher RJ, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 28.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Gelder RN, et al. Pleiotropic effects of cryptochromes 1 and 2 on free-running and light-entrained murine circadian rhythms. J Neurogenet. 2002;16:181–203. doi: 10.1080/01677060215306. [DOI] [PubMed] [Google Scholar]
- 30.Fasano O, Taparowsky E, Fiddes J, Wigler M, Goldfarb M. Sequence and structure of the coding region of the human H-ras-1 gene from T24 bladder carcinoma cells. J Mol Appl Genet. 1983;2:173–180. [PubMed] [Google Scholar]
- 31.Ishizaki K, et al. Increased UV-induced SCEs but normal repair of DNA damage in p53-deficient mouse cells. Int J Cancer. 1994;58:254–257. doi: 10.1002/ijc.2910580218. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 33.Oishi K, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 34.Barnes JW, et al. Requirement of mammalian Timeless for circadian rhythmicity. Science. 2003;302:439–442. doi: 10.1126/science.1086593. [DOI] [PubMed] [Google Scholar]
- 35.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gery S, et al. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 37.Davis S, Mirick DK, Stevens RG. Night shift work, light at night, and risk of breast cancer. J Natl Cancer Inst. 2001;93:1557–1562. doi: 10.1093/jnci/93.20.1557. [DOI] [PubMed] [Google Scholar]
- 38.Schernhammer ES, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses' health study. J Natl Cancer Inst. 2001;93:1563–1568. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 39.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith JJ, et al. Wee1-regulated apoptosis mediated by the crk adaptor protein in Xenopus egg extracts. J Cell Biol. 2000;151:1391–1400. doi: 10.1083/jcb.151.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith JJ, et al. Apoptotic regulation by the Crk adapter protein mediated by interactions with Wee1 and Crm1/exportin. Mol Cell Biol. 2002;22:1412–1423. doi: 10.1128/mcb.22.5.1412-1423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riou L, et al. Differential repair of the two major UV-induced photolesions in trichothiodystrophy fibroblasts. Cancer Res. 2004;64:889–894. doi: 10.1158/0008-5472.can-03-2070. [DOI] [PubMed] [Google Scholar]
- 44.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reardon JT, Sancar A. Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. Meth in Enzymol. 2006;408:189–213. doi: 10.1016/S0076-6879(06)08012-8. [DOI] [PubMed] [Google Scholar]