Abstract
Nuclear factor-erythroid 2 p45-related factor 2 (Nrf2) is the primary transcription factor protecting cells from oxidative stress by regulating cytoprotective genes, including the antioxidant glutathione (GSH) pathway. GSH maintains cellular redox status and affects redox signaling, cell proliferation, and death. GSH homeostasis is regulated by de novo synthesis as well as GSH redox state; previous studies have demonstrated that Nrf2 regulates GSH homeostasis by affecting de novo synthesis. We report that Nrf2 modulates the GSH redox state by regulating glutathione reductase (GSR). In response to oxidants, lungs and embryonic fibroblasts (MEFs) from Nrf2-deficient (Nrf2−/−) mice showed lower levels of GSR mRNA, protein, and enzyme activity relative to wild type (Nrf2+/+). Nrf2−/− MEFs exhibited greater accumulation of glutathione disulfide and cytotoxicity compared to Nrf2+/+ MEFs in response to t-butylhydroquinone, which was rescued by restoring GSR. Microinjection of glutathione disulfide induced greater apoptosis in Nrf2−/− MEFs compared to Nrf2+/+ MEFs. In silico promoter analysis of the GSR gene revealed three putative antioxidant-response elements (ARE1, −44; ARE2, −813; ARE3, −1041). Reporter analysis, site-directed mutagenesis, and chromatin immunoprecipitation assays demonstrated binding of Nrf2 to two AREs distal to the transcription start site. Overall, Nrf2 is critical for maintaining the GSH redox state via transcriptional regulation of GSR and protecting cells against oxidative stress.
Keywords: Nrf2, Oxidative stress, Glutathione, Glutathione reductase, Cigarette smoke, COPD, Emphysema, Free radicals
Because of their function (gas exchange) and structure (large surface area), the lungs are constantly challenged with oxidative insults caused by exogenous air pollutants. Oxidative stress is involved in the pathogenesis of several chronic lung inflammatory diseases, including chronic obstructive pulmonary disease (COPD), asthma, acute respiratory distress syndrome (ARDS), and pulmonary fibrosis. Glutathione (GSH) is a vital antioxidant that regulates the cellular redox status and protects airway epithelial cells from oxidant-induced lung injury and inflammation [1]. Oxidative stress causes perturbations in cellular GSH levels by either affecting the biosynthesis or altering the ratio of intracellular reduced and oxidized forms of glutathione that affect multiple physiological responses. The key functions of GSH include (i) working as a direct antioxidant to aid in the removal of deleterious reactive oxygen species and reactive nitrogen species; (ii) acting as a cofactor for antioxidant enzymes, including glutathione peroxidases, glutathione S-transferases, and glutaredoxins; (iii) recycling of other antioxidants (such as ascorbic acid) [2]; (iv) protecting against inflammatory responses by modulating redox-regulated signal transduction and redox-sensitive transcription factors (NF-κB and AP1) [3]; (v) regulating cell proliferation [4,5]; and (vi) protecting against apoptosis [6,7]. Elevated levels of glutathione disulfide (GSSG) can affect the function of multiple proteins via glutathionylation and cause cell death through lowering reduced GSH levels and increasing accumulation of GSSG [8–10]. The contribution of protection against oxidative stress by regeneration of GSH from GSSG is unclear.
Glutathione reductase (GSR), a homodimeric flavoprotein (50-kDa subunits), regulates cellular GSH homeostasis by catalyzing the reduction of GSSG to GSH using NADPH as a reducing cofactor [11]. Evolutionarily, GSR is well conserved in plants, bacteria, fungi, yeast, and mammals, suggesting that it is important for survival in aerobic environments [11]. GSR has been shown to be critical for cell survival, and mice with suboptimal levels of GSR are more prone to oxidative stress and its associated diseases [12]. However, GSR deficiencies are uncommon among humans, caused primarily by dietary riboflavin deficiencies [13,14] and treatment with the anticancer agent 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU) [15,16].
The basic leucine-zipper transcription factor nuclear factor-erythroid 2 p45-related factor 2 (Nrf2) has been shown to play a vital role in protecting cells from oxidative stress [17]. In response to oxidative stress, Nrf2 dissociates from its cytosolic inhibitor Keap1, translocates to the nucleus, and binds to antioxidant-response elements (AREs) in the promoters of target genes. This leads to transcriptional induction of several cellular defense genes, including glutathione biosynthetic enzymes (glutathione cysteine ligase modifier subunit (GCLM) and glutathione cysteine ligase catalytic subunit (GCLC)), GSH-dependent antioxidant enzymes (glutathione peroxidase 2 (GPX2), glutathione S-transferases (GST), and heme oxygenase-1 (HO-1)) [17]. However, disruption of Nrf2 in mice diminishes or abrogates the induction of these antioxidant genes, indicating their Nrf2-dependent regulation. We and others have shown that Nrf2-defcient (Nrf2−/−) mice show enhanced sensitivity to a variety of lung inflammatory diseases, including cigarette smoke (CS)-induced emphysema [18], allergic asthma [19], endotoxin [20], hyperoxia-induced acute lung injury [5], and bleomycin-induced lung fibrosis [21], in which increased oxidative stress results from impaired adaptive induction of GSH and GSH-dependent enzymes. More recently, we and others have shown that a decline in Nrf2 in the lungs of patients with COPD contributes to pulmonary oxidant/antioxidant imbalance and pathogenesis of lung diseases [22–24]. In contrast, because of a functional mutation in the Keap1 gene, lung cancer tissues show constitutively higher Nrf2-dependent antioxidants, including GSH, which in turn cause therapeutic resistance to chemotherapeutic drugs through attenuation of oxidative stress [25].
Several lines of evidence indicate that Nrf2 plays a key role in the regulation of cellular GSH homeostasis: (i) there is low GSH or a loss of induction of GSH in Nrf2−/− cells and tissues [18,20]; (ii) Nrf2 regulates GSH biosynthesizing enzymes (GCLM, GCLC) [26]; (iii) Nrf2 regulates the cysteine/glutamate exchange transporter that maintains intracellular GSH levels by regulating cysteine influx [27]; and (iv) Nrf2 regulates GPX2 and GST, which use GSH as a cofactor [28]. In addition, we and others have shown that in response to oxidative stress, Nrf2−/− cells and tissues accumulate greater levels of GSSG than wild-type Nrf-2 cells [22]. Because GSR is the key enzyme that mediates the reduction of GSSG and regeneration of GSH, we hypothesized that Nrf2 regulates GSR during oxidative stress. This study elucidates the critical role of GSR in cell survival and molecular regulation of GSR by Nrf2 in response to oxidative stress.
Materials and methods
Animals and care
Nrf2−/− CD-1 (ICR) mice were generated as described [12]. All experimental procedures conducted on the mice were performed in accordance with the standards established by the U.S. Animal Welfare Acts, set forth in the National Institutes of Health guidelines and the Policy and Procedures Manual of the Johns Hopkins University Animal Care and Use Committee.
Mouse exposure to cigarette smoke
Eight-week-old mice were divided into four groups (n=3 per group): I, air control Nrf2+/+ mice; II, experimental Nrf2+/+ mice; III, air control Nrf2−/− mice; and IV, experimental Nrf2−/− mice. Groups I and III were kept in a filtered-air environment, and groups II and IV were subjected to CS according to a previously published protocol [18]. CS exposure was performed by burning 3R4F reference cigarettes (0.73 mg nicotine per cigarette, purchased from the Tobacco Research Institute, University of Kentucky, Lexington, KY, USA) using a smoking machine (Model TE-10; Teague Enterprises, Woodland, CA, USA) as previously described [29]. Six cigarettes were smoked simultaneously, resulting in an exposure level of100 mg/m3. Mice were exposed to this concentration 5 hday for 1 day.
Cell culture
Mouse embryonic fibroblasts (MEFs) were isolated from Nrf2+/+, Nrf2−/−, and Keap1−/− mice as previously described [20] and were cultured in Iscove’s modified Dulbecco’s medium supplemented with 10% FBS and 1% penicillin–streptomycin (Invitrogen, Carlsbad, CA, USA).
Knockdown of GSR by short interfering RNA (siRNA)
Four GSR siRNA duplexes and siControl nontargeting siRNA 1 (SS siRNA) were obtained from Dharmacon Research (Lafayette, CO, USA). First, we identified an siRNA duplex that exhibits selective and maximal silencing of the GSR gene compared to SS siRNA. Briefly, Nrf2+/+ MEFs at 80% confluency were transfected with 20 pmol of siRNA duplexes using Lipofectamine 2000 and OPTI-MEM I reduced-serum medium (Invitrogen) according to the Lipofectamine protocol. Concentrations of siRNAs were chosen on the basis of dose–response studies. Knockdown of the GSR gene was quantified by real-time reverse transcriptase-polymerase chain reaction (RT-PCR) 48 h after transfection. The sequences of the sense strands of the siRNA duplexes were as follows: duplex 1, AGACGAAGCUGUUCAUAAGUU; duplex 2, GACCAUGAUUCCAGAUGUUUU; duplex 3, GACGGGACCCAAAUUCUAAUU; and duplex 4, CGUGAAUGUUGGAUGUGUAUU. Duplex 1 was shown to have the greatest knockdown efficiency (data not shown).
Overexpression of GSR
The vector for GSR overexpression was obtained from Open Biosystems (Huntsville, AL, USA) (Clone ID 6813295). The cDNA insert was digested out of the parent plasmid and ligated into the pUB6-V5-His mammalian expression vector (Invitrogen) with a ubiquitin promoter. After amplification in Escherichia coli and plasmid purification (Qiagen Sciences, Gaithersburg, MD, USA), the plasmid was transfected into Nrf2−/− MEFs using Lipofectamine 2000 (Invitrogen) as described above.
Cytoprotective role of GSR
Microinjection of GSSG and measurement of apoptosis
Nrf2+/+ and Nrf2−/− MEFs were plated at a density of 1×106 cells/ml in Mattek dishes. Approximately 100 cells per dish were injected with vehicle, phosphate-buffered saline (PBS), GSSG, or GSH (Sigma–Aldrich, St. Louis, MO, USA) at a concentration of 50 mM using a FemtoJet microinjector (Eppendorf North America, Westbury, NY, USA). As a control for injection efficiency, cells were also co-injected with Alexa fluorhydrazine 596 (Invitrogen). After the 4-h incubation at 37°C in 5% CO2, the number of apoptotic cells was determined by annexin V Alexa fluor 488 (Invitrogen) staining following the manufacturer’s protocol. Data are expressed as the percentage of apoptotic cells compared to total number of cells microinjected.
tert-Butylhydroquinone-induced cell death
tert-Butylhydroquinone (tBHQ)-induced cytotoxicity was evaluated in (i) Nrf2+/+ and Nrf2−/− MEFs; (ii) Nrf2+/+ MEFs transfected with GSR siRNA or SS siRNA; and (iii) Nrf2−/− MEFs overexpressing GSR or plasmid vector. Briefly, MEFs were seeded in 96-well plates at a density of 1×105 cells/ml and grown overnight. Cells were treated with tBHQ (150 μM) for 4 h and cell death was evaluated using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma) reduction conversion assay [30]. Cell survival was expressed as absorbance relative to that of vehicle-treated controls. For GSR knockdown or overexpression experiments, 48 h after transfection, MEFs were challenged with tBHQ.
Measurement of oxidized and reduced glutathione
Nrf2+/+ and Nrf2−/− MEFs were seeded in 96-well plates at a density of 1×105 cells/ml and were treated with or without BCNU (25 μM), an irreversible inhibitor of GSR, for 16 h. Intracellular levels of total glutathione, GSH, and GSSG were determined using an enzymatic recycling assay as described previously [31].
GSR mRNA expression by quantitative real-time RT-PCR
Total RNA was extracted from lung tissues or cells with TRIzol reagent (Invitrogen) and reverse transcribed using the Superscript First Strand Synthesis system (Invitrogen) as per the manufacturer’s instructions. Quantitative real-time RT-PCR analyses of murine GSR was performed by using Assay on Demand (Cat. No. Mm00439151_m1) primers and probe sets from Applied Biosystems (Foster City, CA, USA). The assay was performed using the ABI 7000 TaqMan system (Applied Biosystems). β-Actin was used for normalization.
Immunoblot analysis of GSR
For immunoblot analysis, 40 μg of total protein was separated by 4–12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to PVDF membrane by semidry blotting. The PVDF membrane was blocked and incubated with polyclonal rabbit anti-GSR antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) (1:1000) or rabbit anti-GAPDH (1:1000), followed by incubation with horseradish peroxidase-conjugated horse anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA), and developed using an ECL chemiluminescence detection system (Amersham Biosciences, Piscataway, NJ, USA).
Immunohistochemical analysis of GSR in mouse lungs
Nrf2+/+ and Nrf2−/− mice were exposed to CS for 5 h. After 24 h, the mice were sacrificed, and the lungs were processed for immunohistochemical analysis as described previously [28]. Briefly, 6-μm-thick lung sections were heated at 95°C for 10 min in 10 mM citrate buffer (pH 6) and allowed to cool for 10 min. The sections were blocked in 10% goat serum for 30 min; endogenous biotin was blocked using an avidin–biotin blocking kit (Zymed, South San Francisco, CA, USA). Sections were then incubated with 1.0 μg/ml rabbit anti-GSR antibody [32] for 18 h at 4°C. Nonimmune rabbit IgG (Jackson ImmunoResearch Laboratories) was used as a negative control. Antibody binding was detected by incubation with biotinylated goat anti-rabbit antibody diluted 1:1000 (Jackson ImmunoResearch Laboratories). Antibody binding was visualized by incubation with avidin–peroxidase conjugate (Vector Laboratories, Burlingame, CA, USA) for 30 min and with DAB (Zymed) for 5 min.
GSR enzyme activity
To measure the enzyme activity of GSR, mice were exposed to CS for 5 h and sacrificed 24 h later. The lungs were excised (n=5 per group) and processed as described previously [18]. GSR activity was measured via NADPH consumption using a spectrophotometer at 340 nm [11].
Identification of AREs in the promoter of GSR
To identify the presence and location of AREs in the GSR (Accession No. 14782) promoter, the 2-kb upstream region from the translation start site was downloaded from the NCBI database (www.ncbi.nlm.nih.gov). The 2-kb sequence was used to search for AREs with the help of Genamics Expression 1.1 software (Hamilton, New Zealand) using the primary ARE core sequence (RTGAYNNNGCR) as the probe. The location of the transcription start site for the mouse GSR gene was determined using Mouse Genome build 33, version 1, of the NCBI database. The promoter sequences of mouse and human homologs of GSR were downloaded from the NCBI database and scanned for the presence of AREs.
Plasmids and mutagenesis
The 5′ flanking region of the mouse GSR promoter region was PCR amplified from genomic DNA isolated from murine blood with high-fidelity Taq polymerase (Applied Biosystems). The isolated PCR product was ligated into pCR2.1 (Invitrogen), and a KpnI–XhoI fragment from this construct was cloned into the pGL3 Basic vector (Promega, Madison, WI, USA). Three deletion constructs (− 1253 to +624, −978 to +624, and −755 to +624) were generated. The primers used for amplification were as follows: AAGTCAAAGTAACGCTGGTGTTGG (ARE1–3 forward), CCCTTTTGTGATCAAGTAGGAGTT (ARE1–2 forward), ACAGGGTACTCTACCAGAAGGAAA (ARE1 forward), and CTGTTACCTAGCACTTTGCCCTTG (reverse primer for all constructs).
Individual AREs identified in the mouse GSR promoter region were PCR amplified from the ARE1–3 constructs in the pGL3 Basic vector using high-fidelity Taq polymerase (Applied Biosystems). The isolated PCR product was ligated into pCR2.1 (Invitrogen), and a KpnI–XhoI fragment from this construct was cloned into the pTAL luciferase reporter vector (BD Biosciences, San Jose, CA, USA). The forward primers used for amplification were as follows: ARE3 forward (−1073), ACTGGTTATTGCCTCACAACGGCA; ARE2 forward (− 857), TTAACTCGGTGATCTTAGCAATCA; and ARE1 forward (− 143), TGCAGGAATCCGAGAAGC. The reverse primers used for amplification were as follows: ARE3 reverse (−997), CCGGCAGAAAAGTTGGTTTCCCTT; ARE2 reverse (−725), CCTTCTTCCTTGATGATCTTGAAC; and ARE1 reverse (+14), ACTTCCGCGCATGGCGCT. Mutated (mu) ARE sequences were generated by using a site-directed mutagenesis kit from Stratagene (La Jolla, CA, USA). Primers containing the mu-ARE sequences (mu-ARE3, TTCTTATGACTTATTAGTATTTAA; mu-ARE2, AAGAAAGCCTTTTGGTCACTGTGA; mu-ARE1, GGCGCGGTTTTTAGTCACGGCGAC) were used for PCR amplification of the mutated GSR promoter, and PCR products were digested with DpnI for 1 h to cleave the wild-type promoter. The mutation in each promoter was verified by sequencing. The NQO1 ARE reporter construct contains a 41-bp rQR-ARE/EpRE inserted into a minimal promoter vector containing a TATA box and an initiator element [33].
DNA transfection and luciferase activity
Cells were transfected at 80% confluence using Lipofectamine 2000 (Invitrogen). Briefly, cells were seeded in 24-well plates at a density of 2×105 cells/ml and grown overnight. The subsequent day, cells were transfected with 200 ng of plasmid DNA and 1 ng of pRL-TK plasmid (Promega). After 48 h, cells were lysed, and Renilla and firefly luciferase activities were measured using the dual luciferase assay kit (Promega) with a luminometer (EG&G Wallac, Gaithersburg, MD, USA). For transfection efficiency, luciferase activity was normalized to Renilla luciferase activity.
Chromatin immunoprecipitation assay
MEFs(Keap−/−, Nrf2+/+, and Nrf2−/−)were harvested and the chromatin immunoprecipitation(CHIP)assay was performed using a commercially available kit (Upstate Biotechnology, Lake Placid, NY, USA). Immunoprecipitates and total chromatin input were reverse cross-linked, DNA was isolated, and 1μl of DNA was used for PCR(35cycles)with primers specific for the individual GSR AREs. The GSR primer sequences were as follows: (1) ARE2 forward, CCATCAAACTTTAACTCGGTGA; reverse GACTTGGGAGATAGAAGGAACG; (2) ARE3 forward, TGAGATTGACTGACACAATGGA; reverse, GATCACAAAAGGGAAACCAACT.
Statistical analysis
Results are presented as the means±SE. Statistical comparisons were performed by paired Student t tests. A value of p < 0.05 was considered statistically significant.
Results
Nrf2-dependent expression of glutathione reductase in mouse lung after acute exposure to CS
The basal levels of mRNA expression of GSR in the lungs were similar in Nrf2+/+ and Nrf2−/− mice as determined by real-time RT-PCR analysis. In response to CS exposure, the mRNA expression of GSR showed an approximately fourfold induction in the lungs of Nrf2+/+ mice compared to air, whereas the lungs of Nrf2−/− mice showed no induction of GSR (Fig. 1A). In corroboration, immunoblot analysis showed no differences in the basal levels of GSR protein in Nrf2+/+ and Nrf2−/− lungs; however, CS exposure elevated GSR protein only in the Nrf2+/+ lungs (Figs. 1B and C). Along the same line, GSR-specific activity also was elevated only in Nrf2+/+ lungs, whereas there was no induction in Nrf2−/− lungs after CS exposure (Fig. 1D). Immunohistochemical analysis of mouse lungs with anti-GSR antibody showed increased staining in airway epithelial cells of CS-exposed Nrf2+/+ mice, whereas very weak or no staining was observed in lung sections of Nrf2+/+ air controls and air- or CS-exposed Nrf2−/− mice (Fig. 1E). Taken together, these results indicate that induction of GSR in mouse lungs during CS exposure is dependent on Nrf2.
Fig. 1.
Nrf2-dependent induction of glutathione reductase in the lungs of mice in response to cigarette smoke-induced oxidative stress. (A) Quantification of mRNA expression of GSR in Nrf2+/+ and Nrf2−/− lungs after CS exposure. Lungs were isolated from mice immediately after 5 h of CS exposure and mRNA expression of GSR was analyzed by real-time RT-PCR. Data are presented as the means±SD; n=3. (B) Protein expression of GSR in Nrf2+/+ and Nrf2−/− lungs after CS exposure. Lungs were isolated from mice immediately after 5 h of CS, and GSR expression was analyzed by Western blot. Immunoblot is a representative of two mouse lungs from three independent analyses. (C) Quantification of GSR protein expression in the lungs of air- and CS-exposed Nrf2+/+and Nrf2−/− mice; n=6 per group. *Significant compared to air controls; p < 0.05. (D) GSR enzyme activity in the lungs of Nrf2+/+ and Nrf2− mice 24 h after CS exposure. Enzyme activity was measured in lung lysates and expressed as nmol of NADPH oxidized/min/mg protein; n=4 per group. (E) Immunohistochemical analysis of GSR protein in the lungs of Nrf2+/+ and Nrf2−/− mice after CS exposure. At 24 h post-CS exposure, lungs were fixed and processed for immunohistochemical analysis using anti-GSR antibody. Intense staining was observed in the epithelial cells lining the small and large airways of CS-exposed Nrf2+/+ mice (E-II), whereas weak or no staining was observed in the lung sections of the CS-exposed Nrf2−/− mice (E-IV) and the air-exposed Nrf2+/+ (E-I) and Nrf2−/− mice (E-III). Original magnification: 20×. Abbreviations are as follows: BV, blood vessel; AW, airway; and AE, airway epithelium. For (A) and (C), relative fold change is abbreviated RFC.
tBHQ treatment induces GSR expression only in Nrf2+/+ MEFs
In support of in vivo data on CS exposure, we measured GSR expression in Nrf2+/+ and Nrf2−/− MEFs after tBHQ (50 μM) treatment. Nrf2+/+ MEFs showed an ~2-fold increase in GSR mRNA and protein expression 16 h after tBHQ treatment compared to vehicle control (DMSO) (Figs. 2A and B). However, tBHQ treatment failed to induce GSR expression in Nrf2−/− MEFs. Consistent with the in vivo data, basal levels of GSR expression were similar in Nrf2+/+ and Nrf2−/− MEFs (Fig. 2C). Disruption of Keap1 in MEF cells led to increased basal Nrf2 activity, with Keap−/− MEF cells displaying an ~1.9-fold increase in basal GSR mRNA expression relative to Nrf2+/+ cells (Fig. 2D).
Fig. 2.
Activation of Nrf2 by tBHQ treatment and disruption of Keap1 induces GSR expression. (A) Expression of GSR mRNA in Nrf2+/+ and Nrf2−/− MEFs treated with tBHQ. MEFs were challenged with either vehicle (DMSO) or tBHQ (50 μM) for 16 h. After challenge, mRNA expression was analyzed by RT-PCR. The mRNA levels were normalized to GAPDH endogenous control; n=3. (B) Protein expression of GSR in Nrf2+/+ and Nrf2−/− MEFs treated with tBHQ. MEFs were challenged with tBHQ (50 μM), and 16 h later, GSR protein expression was analyzed by immunoblot. Immunoblot is representative of two biological duplicates of three independent analyses. (C) Quantification of GSR protein expression in the MEFs after tBHQ treatment; n=6. (D) Basal expression of GSR mRNA in Nrf2+/+, Nrf2−/−, and Keap−/− MEFs as analyzed by real-time RT-PCR. Data are represented as fold change relative (RFC) to Nrf2+/+ cells; n=3. *Significant compared to Nrf2+/+ or vehicle; p < 0.05.
Oxidized glutathione induces greater apoptosis in Nrf2-deficient cells
To determine the role of Nrf2 in conferring cytoprotection against intracellular GSSG, Nrf2+/+ and Nrf2−/− MEFs were microinjected with GSSG (50 μM), reduced glutathione (50 μM), or PBS. Cells with an approximate intracellular volume of 1 pl (picoliter) were injected with 1 fl (femtoliter) of each treatment. Approximately 100 cells were injected with each treatment, and apoptosis was determined by annexin V binding 1.5 h after injection. MEFs injected with the three treatments on a single plate are shown in Fig. 3A. PBS or GSH microinjection caused no cell death in both genotypes. However, GSSG induced greater apoptosis in Nrf2−/− MEFs compared to wild-type MEFs (Fig. 3B). These results indicate a greater ability of Nrf2+/+ MEFs to detoxify GSSG compared to Nrf2−/− MEFs.
Fig. 3.
Oxidized glutathione induces apoptosis in Nrf2+/+ and Nrf2−/− MEFs. Approximately 100 MEFs per genotype were microinjected with GSSG (50 μM), GSH (50 μM), or PBS solution. Apoptosis was analyzed by annexin V binding 90 min after microinjection. (A) Photomicrographs illustrating annexin V staining in cells microinjected with GSSG compared to no staining seen in cells microinjected with GSH and PBS. (B) Percentage of apoptotic cells in Nrf2+/+ and Nrf2−/− MEFs 90 min after GSSG microinjection. Values presented are percentage annexin V-positive cells of the total number of injected cells. (C) Transfection of GSR overexpression vector increased GSR mRNA expression in Nrf2−/− MEFs 48 h posttransfection. *Significant relative to Nrf2+/+ vehicle, and †significant relative to Nrf2−/− MEF cells transfected with empty vector (p < 0.05). (D) Overexpression of GSR rescues Nrf2−/− MEFs from tBHQ-induced cytotoxicity. Results are presented as percentage cell death relative to untreated Nrf2+/+ MEFs; n=3. *Significant relative to control (p < 0.05).
Restoration of GSR rescues Nrf2−/− MEFs from tBHQ-induced cell death
To determine whether restoration of GSR protects Nrf2−/− MEFs from tBHQ-induced cytotoxicity, Nrf2−/− MEFs were transfected with the GSR overexpression vector. After 48 h of transfection, an ~10-fold increase in the GSR mRNA expression in Nrf2−/− MEFs compared to vector alone was noted (Fig. 3C). To determine the impact on tBHQ-induced cytotoxicity, GSR-overexpressing Nrf2−/− MEFs were treated with tBHQ (150 μM), and cell death was assessed after 6 h. Overexpression of GSR in Nrf2−/− MEFs significantly attenuated tBHQ-induced cytotoxicity compared to Nrf2−/− MEFs transfected with vector alone (Fig. 3D).
Glutathione reductase maintains GSH redox state and is critical for cell survival
Next, we investigated whether knockdown of GSR in Nrf2+/+ MEFs sensitizes cells to tBHQ-induced cytotoxicity. MEFs transfected with GSR siRNA showed >90 and 65% reduction mRNA levels and enzyme activity of GSR, respectively, compared to mock and scrambled siRNA (ssRNA) (Figs. 4A and B). MEFs transfected with GSR siRNA and ssRNA were challenged with tBHQ (150 μM), and cell death was measured by MTT assay after 6 h. The tBHQ treatment did not induce any significant cell death in ssRNA-transfected cells; however, ~40% increased cell death was observed in cells transfected with GSR siRNA compared to controls (Fig. 4C). No cell death was observed in vehicle-treated cells transfected with SS siRNA or GSR siRNA.
Fig. 4.
Glutathione reductase maintains GSH redox state and is critical for cell survival. (A) GSR mRNA expression in Nrf2+/+ MEFs 48 h after GSR siRNA, SS siRNA, and mock transfection. Data are expressed as the relative fold change (RFC)±SD; n=3. (B) Basal GSR enzyme activity in MEFs 48 h after transfection with SS siRNA and GSR siRNA. Enzyme activity was measured in cell lysates and is expressed as nmol of NADPH oxidized/min/mg protein; n=3. (C) tBHQ-induced cell death in MEFs transfected with GSR siRNA. After 48 h of GSR siRNA transfection, MEFs were treated with tBHQ (150 μM) for 4 h, and cell death was analyzed by MTT assay; n=3. (D) Levels of total GSH in MEFs 16 h after BCNU, a potent inhibitor of GSR treatment; n=3. (E) Levels of reduced GSH and GSSG are expressed as a percentage of total GSH. Significant compared to vehicle control at p<0.05. (F) Levels of total, oxidized, and reduced glutathione in Nrf2+/+ and Nrf2−/− MEFs 24 h after treatment with tBHQ (100 μM). For each treatment and genotype, reduced GSH and GSSG levels are presented as relative percentages of total glutathione in control Nrf2+/+ vehicle-treated cells (RTC).
GSR regulates GSH homeostasis
To determine the role of GSR in the maintenance of GSH homeostasis, wild-type MEFs were treated with BCNU, a selective inhibitor of GSR [34,35], and 16 h later, GSH and GSSG were assessed. Consistent with previous reports, BCNU treatment significantly elevated total glutathione compared to vehicle treatment (Fig. 4D); however, 75% of total glutathione was GSSG in BCNU-treated cells, whereas only 5% of total glutathione was GSSG in vehicle-treated cells (Fig. 4E). An attempt was made to assess cell viability via an MTT assay after BCNU and tBHQ treatment; however, treatment conditions resulted in total cell death. These results emphasize the role of GSR in maintaining GSH/GSSG homeostasis and protection against oxidative stress-induced cell death.
Nrf2−/− cells show enhanced accumulation of GSSG during oxidative stress
Constitutively, Nrf2+/+ MEFs showed significantly higher levels of total glutathione compared to Nrf2−/− MEFs (Fig. 4F). Interestingly, the percentage of the total glutathione that was GSSG was greater in Nrf2−/− MEFs compared to Nrf2+/+ MEFs. In response to tBHQ treatment, there was a significant decrease in total glutathione in MEFs of both genotypes. However, Nrf2−/− MEFs showed greater accumulation of GSSG compared to Nrf2+/+ MEFs after tBHQ treatment (Fig. 4E). Taken together, these results demonstrate that Nrf2 deficiency results in greater accumulation of cellular oxidized glutathione due to lower expression of GSR.
Nrf2 regulates glutathione gene expression through the ARE
Detailed in silico analysis of the 2-kb promoter region 5′ upstream of the translational start site of the mouse GSR genomic locus was performed using the core ARE sequence RTGAYNNNGCR with the help of Genamics Expression 1.1 software as previously described [28]. Three putative AREs in the murine GSR promoter were indicated: ARE3 at −1041 to −1030, ARE2 at −813 to −802, and ARE1 at −44 to −33. Sequences of ARE2 and ARE1 were in reverse orientation, whereas ARE3 showed a forward orientation. Sequence homology between mouse and human GSR promoters was minimal (only 60%) (Fig. 5A); however, 90% sequence homology was observed between mouse and human GSR cDNA (data not shown). All three putative AREs in the GSR promoter showed high sequence homology to the NQO1 ARE (Fig. 5B).
Fig. 5.
Nrf2 mediates transcriptional regulation of the GSR gene via the ARE. Functional analysis of three AREs in the GSR promoter by transient transfection in Nrf2−/−, Nrf2+/+, and Keap−/− MEFs. (A) A 2-kb portion 5′ upstream of the translation start site of the GSR gene was analyzed for the presence of AREs. The three AREs identified are highlighted in red with orientation indicated by the black arrows. The transcription start site is indicated in yellow. (B) The sequence alignment of the individual AREs and the NQO1 ARE is shown along with the consensus sequence. (C) Fragments of the GSR promoter containing all three AREs and deletion constructs containing ARE1 and ARE2 or only ARE1 were cloned into the pGL3 Basic luciferase reporter vector. These constructs were transiently transfected into Nrf2−/− and Keap−/− MEFs and luciferase activity was recorded after 48 h. (D) The full-length promoter region and individual AREs were cloned into the pTAL luciferase reporter vector with minimal promoter (ARE3, −1703 to −997; ARE2, −857 to −725; and ARE1, −143 to +14). The putative AREs in individual constructs were subjected to site-directed mutagenesis using the primers described under Materials and methods. These constructs (individual and mu-AREs) were transiently transfected into Nrf2−/− MEFs (no Nrf2 activity) and Keap−/− MEFs (high Nrf2 activity) and luciferase activity was measured after 48 h. Luciferase activity was normalized by measuring the Renilla luciferase activity from a cotransfected reporter vector. Values are means±SE from three different experiments (p<0.05). *Significant compared to Nrf2−/−. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To investigate the functionality of these putative AREs in GSR promoter and Nrf2-dependent regulation, we cloned the 2-kb portion 5′ upstream of the translation start site of the GSR gene into the pGL3 Basic luciferase reporter vector. Reporter constructs containing all three putative AREs, two AREs, and one ARE were cloned into the pGL3 Basic vector. Two deletion constructs (−1253 to +624 (ARE1–3), −978 to +624 (ARE1–2)) exhibited ~160-fold higher luciferase activity compared with the pGL3 Basic vector. The smallest clone containing the ARE1 sequence (−755 to +624 (ARE1)) overlapping with the promoter demonstrated ~40-fold higher reporter activity (Fig. 5C). The presence of all three AREs or two AREs (ARE2 and ARE1) resulted in a 4-fold higher reporter activity compared to ARE1 alone. All three reporter constructs containing all three putative AREs exhibited over 3- to 20-fold induction in Keap−/− cells (high Nrf2 activity) compared with Nrf2−/− cells (no Nrf2 activity). These data strongly suggest that ARE sequences in the GSR promoter are active and function as enhancers.
To dissect the functionalities of individual ARE enhancer sequences, we cloned specific regions of the promoter-containing individual AREs into the pTAL luciferase vector with minimal promoter (ARE3, −1703 to −997; ARE2 −857 to −725; and ARE1, −143 to +14) and transiently transfected them into Nrf2−/− and Keap−/− MEFs. The two distal ARE reporter constructs (ARE3 and ARE2) showed significant induction of luciferase activity in Keap−/− MEFs, whereas no induction was observed in Nrf2−/− MEFs. ARE3 showed the highest luciferase activity, followed by ARE2, whereas ARE1 showed no significant activity (Fig. 5D). On the other hand, transfection of individual mu-ARE reporter plasmids in which the consensus ARE was mutated failed to induce luciferase activity in Keap−/− MEFs, and it was comparable to that in Nrf2−/− cells and or vector alone, indicating the specificity of the “ARE motifs” in the induction of reporter activity (Fig. 5D). Taken together, these experiments demonstrate the functionality of the two distal AREs (ARE2 and ARE3) identified in the GSR promoter and the regulatory role of Nrf2 in modulating GSR gene expression.
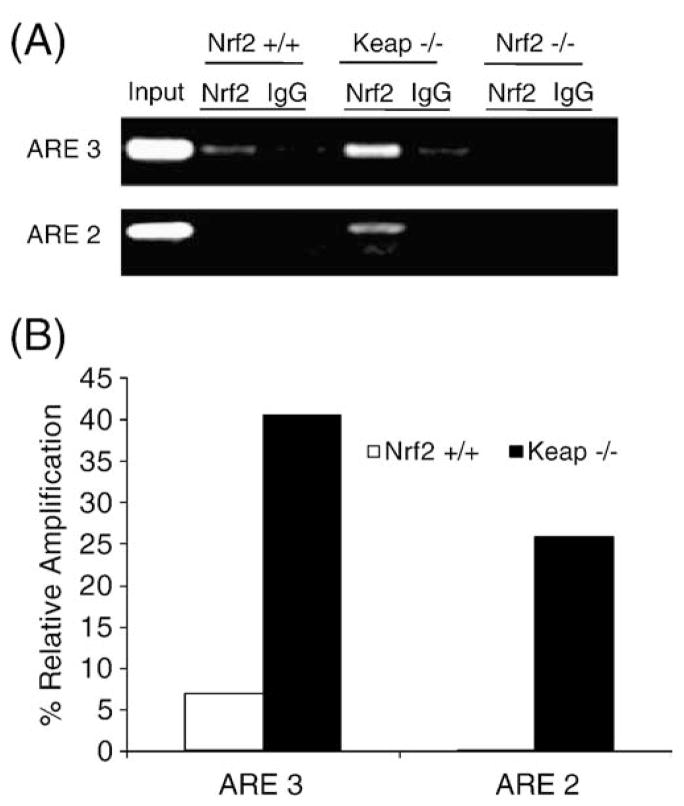
Nrf2 binds to ARE3 and ARE2 in the promoter of the GSR gene
Next, we analyzed the constitutive Nrf2 binding to ARE3 and ARE2 by CHIP assay in Nrf2+/+, Nrf2−/−, and Keap−/− MEFs. A CHIP assay was performed using a commercially available kit (Upstate Biotechnology). Immunoprecipitates and total chromatin input were reverse cross-linked, DNA was isolated, and 1 μl of DNA was used for PCR (35 cycles) with primers specific for the individual GSR AREs present in the promoter. An anti-Nrf2 antibody was used to confirm the binding of Nrf2 to the GSR promoter. Activation of Nrf2 by genetic deletion of Keap1 enhanced the recruitment of Nrf2 to AREs in the GSR promoter as demonstrated by enhanced amplification, with ARE3 showing the highest level of binding (Fig. 6A). Nrf2 was present in undetectable levels in Nrf2−/− MEFs, and no amplification was detected using primers for the GSR AREs. Nonspecific antibody was used as a negative control in these assays. Quantification of CHIP amplification was performed relative to input (Fig. 6B).
Fig. 6.
CHIP analysis demonstrating Nrf2 binding to GSR AREs. (A) Nuclear extracts from Nrf2−/−, Nrf2+/+, and Keap−/− MEFs were used to assess the Nrf2 binding activity to ARE3 and ARE2. (A) CHIP assay was performed with Nrf2−/−, Nrf2+/+, and Keap−/− MEFs using anti-Nrf2 antibody and rabbit IgG1. (B) Quantification of CHIP assay results ((GSR promoter binding-antibody)/input ratio). Quantification was performed on representative CHIP data from n=3 independent experiments.
Discussion
Through the use of Nrf2-disrupted models (cells and mice), our laboratory and others have previously reported that Nrf2 protects from cell death induced by multiple oxidants (H2O2, tBHQ, cigarette smoke, hyperoxia, and anticancer drugs) mainly by alleviating cellular ROS levels [17]. Nrf2 positively regulates antioxidant and electrophile detoxification enzymes (NQO1, GSTs, and GPX2) as well as enzymes that directly regulate levels of glutathione (GCLM, GCLC). Because electrophilic detoxification and glutathione homeostasis are coordinately regulated by Nrf2, it has been difficult to determine the contribution of either pathway in redox homeostasis. Here we report for the first time that Nrf2-dependent regulation of GSH redox status by GSR-mediated recycling of oxidized glutathione is critical for cell survival during oxidative stress, independent of the induction of GSH biosynthetic enzymes. The major findings of this study are: (i) a greater accumulation of GSSG in Nrf2−/− cells in response to tBHQ-induced oxidative stress; (ii) an enhanced cytotoxicity in Nrf2−/− cells following microinjection of GSSG; and (iii) protection from tBHQ-induced cytotoxicity after restoration of GSR in Nrf2−/− cells. Thus, minimizing the GSSG level in the cells through GSR-mediated recycling is a critical factor that determines survival during oxidative stress.
Multiple inflammatory disorders including COPD, ARDS, Alzheimer disease, Parkinson disease, liver disease, heart attack, stroke, and diabetes, as well as HIV infection and AIDS, are associated with elevated GSSG and reduced GSH levels. GSR is the key enzyme that maintains the GSH redox state by converting GSSG to GSH and thus may play a vital role in protecting against oxidative pathologies in these diseases. Chemical inhibition of GSR has been demonstrated to significantly impair the survival of mice in response to hyperoxia [36]. In contrast, over-expression of GSR has been reported to extend the survival of Drosophila by attenuating oxidative damage in response to hyperoxia [37]. Over-expression of GSR in macrophages has been shown to protect from inflammatory diseases such as atherosclerosis [38].
Recent studies have indicated that oxidant-induced alterations in the cellular GSH-to-GSSG ratio in favor of GSSG trigger apoptosis independent of ROS and GSH levels, suggesting that GSSG alone is capable of inducing apoptosis [39,40]. Our study also demonstrates that elevation of intracellular GSSG alone by microinjection is adequate to induce apoptosis (Fig. 3). GSSG accumulation can modulate the mitochondrial membrane potential, which can trigger the release of cytochrome c and mediate an apoptotic signaling cascade [8]. Cellular GSSG levels can be elevated during oxidative stress mainly by two mechanisms: first, direct interaction of oxidants with GSH, causing depletion of cellular GSH, and second, differential expression of the GSH-metabolizing enzymes glutathione peroxidases and GSR. Glutathione peroxidases (GPX) are major cellular antioxidative enzymes that protect from oxidative damage and cytotoxicity caused by hydroperoxides (H2O2 and lipid hydroperoxides). However, GPX-mediated reduction of hydroperoxides results in enhanced GSSG formation through oxidation of GSH. Studies have shown that differential expression of GPX2 and GSR greatly determines cell-specific GSH redox status as well as sensitivity to oxidants [12]. Cadmium, at similar concentrations, induced a greater decrease in GSH/GSSG in C6 glioma cells compared to HepG2 cells. In HepG2 cells the GSR activity was almost threefold higher than the GPX2 activity, whereas in C6 glioma cells the GPX2 activity was greater than the GSR activity [12]. These findings highlight the significance of the coexpression of GSR and GPX in determining sensitivity to oxidant-induced cytotoxicity.
Recently we reported that Nrf2 regulates GPX2, one of the major isoforms of glutathione peroxidase, in lung cells as a response to oxidant or phytochemical treatment [28]. However, it was unclear whether other Nrf2-dependent antioxidative enzymes would protect from the high cellular levels of GSSG generated by GPX2 activity during oxidative stress. The major findings of this study in conjunction with previously published GPX2 data demonstrate that GSR is a key antioxidant gene by which Nrf2 mediates cytoprotection during oxidative stress.
Nrf2 regulates its target gene transcripts by directly binding to a cis-enhancer element referred to as an antioxidant response element that functions in both forward and reverse orientations. In silico analysis of the GSR promoter revealed three potential AREs within 2 kb upstream of the translation start point. The sequences of all three AREs in the GSR promoter were similar to the prototypical AREs identified in Nrf2-regulated antioxidants such as NQO1 and HO-1. A luciferase reporter construct with two distal AREs (ARE2 and ARE3) showed significant induction of luciferase activity only in Keap−/− and not in Nrf2−/− MEFs. Similarly, individual AREs cloned in luciferase reporter constructs with a minimal flanking promoter also showed significant induction of luciferase activity in Keap−/− MEFs, whereas activity in Nrf2−/− MEFs was equivalent to vector control. Mutation in the core sequence of the individual AREs completely abolished the luciferase activity in Keap−/− and Nrf2+/+ MEFs. Similarly, CHIP analysis revealed greater Nrf2 binding activity at ARE2 and ARE3 in Keap−/−MEFs. However, nuclear extracts from Nrf2−/− and Nrf2+/+ showed poor Nrf2 DNA binding activity. These results corroborate the findings that Nrf2 upregulates expression of GSR (mRNA and protein) under inducible but not basal conditions, with induction of GSR being observed in Nrf2+/+ lungs and MEFs only in response to oxidative stress. Taken together, these results suggest that the two distal AREs in the GSR promoter are functional to a variable degree, and Nrf2 regulates inducible but not basal expression of GSR via these AREs.
In conclusion, we report Nrf2-ARE-mediated regulation of GSR induction during oxidative stress. In addition to GSH biosynthesis, Nrf2-dependent regulation of the cellular GSH redox state through GSR is vital for protecting against oxidative stress-induced cellular redox signal perturbations and cell death in lungs and other organs.
Acknowledgments
The authors thank Dr. Thomas W. Kensler for critical review of the manuscript, Dr. Masayuki Yamamoto for sharing the Nrf2−/− mice, and Drs. Michael A. Trush and James P. Kehrer for discussion of chemical inhibitors of GSR. This work was supported by NIH Grant HL081205, NHLBI SCCOR Grant P50HL084945, FAMRI, a Maryland Cigarette Restitution Fund research grant, GM079239, and NIEHS Center Grant ES03819. C.J.H. was supported by NIEHS Training Grant ES07141.
References
- 1.Rahman I, Biswas SK, Jimenez LA, Torres M, Forman HJ. Glutathione, stress responses, and redox signaling in lung inflammation. Antioxid Redox Signaling. 2005;7:42–59. doi: 10.1089/ars.2005.7.42. [DOI] [PubMed] [Google Scholar]
- 2.May JM, Qu ZC, Whitesell RR, Cobb CE. Ascorbate recycling in human erythrocytes: role of GSH in reducing dehydroascorbate. Free Radic Biol Med. 1996;20:543–551. doi: 10.1016/0891-5849(95)02130-2. [DOI] [PubMed] [Google Scholar]
- 3.Rahman I. Regulation of nuclear factor-kappa B, activator protein-1, and glutathione levels by tumor necrosis factor-alpha and dexamethasone in alveolar epithelial cells. Biochem Pharmacol. 2000;60:1041–1049. doi: 10.1016/s0006-2952(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 4.Markovic J, Borras C, Ortega A, Sastre J, Vina J, Pallardo FV. Glutathione is recruited into the nucleus in early phases of cell proliferation. J Biol Chem. 2007;282:20416–20424. doi: 10.1074/jbc.M609582200. [DOI] [PubMed] [Google Scholar]
- 5.Reddy NM, Kleeberger SR, Cho HY, Yamamoto M, Kensler TW, Biswal S, Reddy SP. Deficiency in Nrf2–GSH signaling impairs type II cell growth and enhances sensitivity to oxidants. Am J Respir Cell Mol Biol. 2007;37:3–8. doi: 10.1165/rcmb.2007-0004RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong JS, Steinauer KK, Hornung B, Irish JM, Lecane P, Birrell GW, Peehl DM, Knox SJ. Role of glutathione depletion and reactive oxygen species generation in apoptotic signaling in a human B lymphoma cell line. Cell Death Differ. 2002;9:252–263. doi: 10.1038/sj.cdd.4400959. [DOI] [PubMed] [Google Scholar]
- 7.Ghibelli L, Fanelli C, Rotilio G, Lafavia E, Coppola S, Colussi C, Civitareale P, Ciriolo MR. Rescue of cells from apoptosis by inhibition of active GSH extrusion. FASEB J. 1998;12:479–486. doi: 10.1096/fasebj.12.6.479. [DOI] [PubMed] [Google Scholar]
- 8.Filomeni G, Rotilio G, Ciriolo MR. Glutathione disulfide induces apoptosis in U937 cells by a redox-mediated p38 MAP kinase pathway. FASEB J. 2003;17:64–66. doi: 10.1096/fj.02-0105fje. [DOI] [PubMed] [Google Scholar]
- 9.Yoon HS, Lee IA, Lee H, Lee BH, Jo J. Overexpression of a eukaryotic glutathione reductase gene from Brassica campestris improved resistance to oxidative stress in Escherichia coli. Biochem Biophys Res Commun. 2005;326:618–623. doi: 10.1016/j.bbrc.2004.11.095. [DOI] [PubMed] [Google Scholar]
- 10.Walther UI, Czermak A, Muckter H, Walther SC, Fichtl B. Decreased GSSG reductase activity enhances cellular zinc toxicity in three human lung cell lines. Arch Toxicol. 2003;77:131–137. doi: 10.1007/s00204-002-0421-z. [DOI] [PubMed] [Google Scholar]
- 11.Carlberg I, Mannervik B. Glutathione reductase. Methods Enzymol. 1985;113:484–490. doi: 10.1016/s0076-6879(85)13062-4. [DOI] [PubMed] [Google Scholar]
- 12.Yang MS, Chan HW, Yu LC. Glutathione peroxidase and glutathione reductase activities are partially responsible for determining the susceptibility of cells to oxidative stress. Toxicology. 2006;226:126–130. doi: 10.1016/j.tox.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 13.el-Hazmi MA, Warsy AS. Glutathione reductase in the south-western province of Saudi Arabia—genetic variation vs. acquired deficiency. Haematologia (Budapest) 1989;22:37–42. [PubMed] [Google Scholar]
- 14.el-Hazmi MA, Warsy AS. Riboflavin status in Saudi Arabia—a comparative study in different regions. Trop Geogr Med. 1989;41:22–25. [PubMed] [Google Scholar]
- 15.Frischer H, Ahmad T. Severe generalized glutathione reductase deficiency after antitumor chemotherapy with BCNU [1,3-bis(chloroethyl)-1-nitrosourea] J Lab Clin Med. 1977;89:1080–1091. [PubMed] [Google Scholar]
- 16.Frischer H. Erythrocytic glutathione reductase deficiency in a hospital population in the United States. Am J Hematol. 1977;2:327–334. doi: 10.1002/ajh.2830020402. [DOI] [PubMed] [Google Scholar]
- 17.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1–Nrf2–ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 18.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, Biswal S. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW, Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. The transcription factor NRF2 protects against pulmonary fibrosis. FASEB J. 2004;18:1258–1260. doi: 10.1096/fj.03-1127fje. [DOI] [PubMed] [Google Scholar]
- 22.Goven D, Boutten A, Lecon-Malas V, Marchal-Somme J, Amara N, Crestani B, Fournier M, Leseche G, Soler P, Boczkowski J, Bonay M. Altered Nrf2/Keap1–Bach1 equilibrium in pulmonary emphysema. Thorax. 2008;63:916–924. doi: 10.1136/thx.2007.091181. [DOI] [PubMed] [Google Scholar]
- 23.Malhotra D, Thimmulappa R, Navas-Acien A, Sandford A, Elliott M, Singh A, Chen L, Zhuang X, Hogg J, Pare P, Tuder RM, Biswal S. Decline in NRF2 regulated antioxidants in COPD lungs due to loss of its positive regulator DJ-1. Am J Respir Crit Care Med. 2008;178:592–604. doi: 10.1164/rccm.200803-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Suzuki M, Betsuyaku T, Ito Y, Nagai K, Nasuhara Y, Kaga K, Kondo S, Nishimura M. Downregulated NF-E2-related factor 2 in pulmonary macrophages of aged smokers and COPD patients. Am J Respir Cell Mol Biol. 2008;39:673–682. doi: 10.1165/rcmb.2007-0424OC. [DOI] [PubMed] [Google Scholar]
- 25.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson AM, Nevarea Z, Gipp JJ, Mulcahy RT. Identification of a variant antioxidant response element in the promoter of the human glutamate–cysteine ligase modifier subunit gene: revision of the ARE consensus sequence. J Biol Chem. 2002;277:30730–30737. doi: 10.1074/jbc.M205225200. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki H, Sato H, Kuriyama-Matsumura K, Sato K, Maebara K, Wang H, Tamba M, Itoh K, Yamamoto M, Bannai S. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J Biol Chem. 2002;277:44765–44771. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]
- 28.Singh A, Rangasamy T, Thimmulappa RK, Lee H, Osburn WO, Brigelius-Flohe R, Kensler TW, Yamamoto M, Biswal S. Glutathione peroxidase 2, the major cigarette smoke-inducible isoform of GPX in lungs, is regulated by Nrf2. Am J Respir Cell Mol Biol. 2006;35:639–650. doi: 10.1165/rcmb.2005-0325OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witschi H, Espiritu I, Maronpot RR, Pinkerton KE, Jones AD. The carcinogenic potential of the gas phase of environmental tobacco smoke. Carcinogenesis. 1997;18:2035–2042. doi: 10.1093/carcin/18.11.2035. [DOI] [PubMed] [Google Scholar]
- 30.Scudiero DA, Shoemaker RH, Paull KD, Monks A, Tierney S, Nofziger TH, Currens MJ, Seniff D, Boyd MR. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988;48:4827–4833. [PubMed] [Google Scholar]
- 31.Rahman I, Kode A, Biswas SK. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc. 2006;1:3159–3165. doi: 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko T, Iuchi Y, Kawachiya S, Fujii T, Saito H, Kurachi H, Fujii J. Alteration of glutathione reductase expression in the female reproductive organs during the estrous cycle. Biol Reprod. 2001;65:1410–1416. doi: 10.1095/biolreprod65.5.1410. [DOI] [PubMed] [Google Scholar]
- 33.Tirumalai R, Rajesh Kumar T, Mai KH, Biswal S. Acrolein causes transcriptional induction of phase II genes by activation of Nrf2 in human lung type II epithelial (A549) cells. Toxicol Lett. 2002;132:27–36. doi: 10.1016/s0378-4274(02)00055-3. [DOI] [PubMed] [Google Scholar]
- 34.Kehrer JP. The effect of BCNU (carmustine) on tissue glutathione reductase activity. Toxicol Lett. 1983;17:63–68. doi: 10.1016/0378-4274(83)90036-x. [DOI] [PubMed] [Google Scholar]
- 35.Shinohara K, Tanaka KR. Mechanism of inhibition of red blood cell glutathione reductase activity by BCNU (1,3-bis(2-chloroethyl)-1-nitrosourea) Clin Chim Acta. 1979;92:147–152. doi: 10.1016/0009-8981(79)90108-6. [DOI] [PubMed] [Google Scholar]
- 36.Kehrer JP, Paraidathathu T. Enhanced oxygen toxicity following treatment with 1,3-bis(2-chloroethyl)-1-nitrosourea. Fundam Appl Toxicol. 1984;4:760–767. doi: 10.1016/0272-0590(84)90097-6. [DOI] [PubMed] [Google Scholar]
- 37.Mockett RJ, Sohal RS, Orr WC. Overexpression of glutathione reductase extends survival in transgenic Drosophila melanogaster under hyperoxia but not normoxia. FASEB J. 1999;13:1733–1742. doi: 10.1096/fasebj.13.13.1733. [DOI] [PubMed] [Google Scholar]
- 38.Qiao M, Kisgati M, Cholewa JM, Zhu W, Smart EJ, Sulistio MS, Asmis R. Increased expression of glutathione reductase in macrophages decreases atherosclerotic lesion formation in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2007;27:1375–1382. doi: 10.1161/ATVBAHA.107.142109. [DOI] [PubMed] [Google Scholar]
- 39.Pias EK, Aw TY. Early redox imbalance mediates hydroperoxide-induced apoptosis in mitotic competent undifferentiated PC-12 cells. Cell Death Differ. 2002;9:1007–1016. doi: 10.1038/sj.cdd.4401064. [DOI] [PubMed] [Google Scholar]
- 40.Pias EK, Aw TY. Apoptosis in mitotic competent undifferentiated cells is induced by cellular redox imbalance independent of reactive oxygen species production. FASEB J. 2002;16:781–790. doi: 10.1096/fj.01-0784com. [DOI] [PubMed] [Google Scholar]