Abstract
Background
The hypothesis of the present study was that molecular mechanisms differ markedly when mediating ischemic preconditioning induced by repetitive episodes of ischemia versus classic first- or second-window preconditioning.
Methods and Results
To test this, chronically instrumented conscious pigs were subjected to either repetitive coronary stenosis (RCS) or a traditional protocol of second-window ischemic preconditioning (SWIPC). Lethal ischemia, induced by 60 minutes of coronary artery occlusion followed by reperfusion, resulted in an infarct size/area at risk of 6±3% after RCS and 16±3% after SWIPC (both groups P<0.05, less than shams 42±4%). Two molecular signatures of SWIPC, the increased expression of the inducible isoform of nitric oxide synthase and the translocation of protein kinase Cε to the plasma membrane, were observed with SWIPC but not with RCS. Confirming this, pretreatment with a nitric oxide synthase inhibitor prevented the protection conferred by SWIPC but not by RCS. Microarray analysis revealed a qualitatively different genomic profile of cardioprotection between ischemic preconditioning induced by RCS and that induced by SWIPC. The number of genes significantly regulated was greater in RCS (5739) than in SWIPC (2394) animals. Of the 5739 genes regulated in RCS, only 31% were also regulated in SWIPC. Broad categories of genes induced by RCS but not SWIPC included those involved in autophagy, endoplasmic reticulum stress, and mitochondrial oxidative metabolism. The upregulation of these pathways was confirmed by Western blotting.
Conclusions
RCS induces cardioprotection against lethal myocardial ischemia that is at least as powerful as traditional ischemic preconditioning but is mediated through radically different mechanisms.
Keywords: myocardial infarction, ischemia, nitric oxide
Ischemic preconditioning (IPC), discovered 20 years ago,1 is the most powerful intervention known to protect myocardium and has resulted in one of the most intensive and prolific areas of investigation, yet it has proved difficult to translate the knowledge obtained into clinical therapy. The vast majority of experimental studies in IPC have used a brief episodes of the IPC stimulus on a background of a normal heart with normal coronary arteries, ie, virgin ischemia. Our hypothesis is that the molecular mechanisms that mediate IPC are radically different in the setting of chronic ischemia, more akin to the situation in patients with coronary artery disease.
To test this hypothesis, we compared the effects of repetitive ischemia, induced by 6 episodes of 90-minute coronary stenosis and 12-hour reperfusion, with a more traditional protocol of IPC, ie, delayed or second-window IPC (SWIPC). Our first goal was to confirm that classic mediators of SWIPC, eg, inducible nitric oxide synthase (iNOS) and protein kinase C (PKC)ε,2–7 were upregulated and, accordingly, that the protection afforded by SWIPC could be abolished by pretreatment with a nitric oxide (NO) synthase inhibitor, N-nitro-l-arginine (L-NNA).2,3,5,7 Failure to uncover these mechanisms with repetitive coronary stenosis (RCS), confirmed by the inability to abolish the protection afforded by RCS with L-NNA, led us to investigate the molecular mechanisms that mediate RCS. This was accomplished by microarray analysis of the IPC myocardium after SWIPC and RCS. We then verified the novel mechanisms by Western blotting, which confirmed that several mechanisms that may mediate RCS were not upregulated in SWIPC.
Methods
Animal Models
Experiments were conducted in female pigs weighing 18 to 22 kg. After general anesthesia with isoflurane (1.5 to 2.0 vol% in oxygen), a left thoracotomy was performed at the fifth intercostal space. A miniature solid state left ventricular (LV) pressure gauge was implanted in the LV through the apex for measurements of LV pressure and rate of change of LV pressure (LV dP/dt). Tygon catheters (Saint Gobain Performance Plastics, Taunton, Mass) were implanted in the descending aorta and left atrium for measurements of their respective pressures. Two pairs of ultrasonic dimension crystals were implanted in the anterior and posterior wall, transmurally, for measurements of nonischemic and potentially ischemic zone wall thickness. A hydraulic occluder was implanted around the left circumflex coronary artery to induce coronary artery occlusion (CAO) or stenosis, and a Transonic blood flow probe (Transonic Systems, Inc, Ithaca, NY) was implanted proximal to the occluder. Five groups of pigs were used for the physiology study, ie, control (n=5), SWIPC (n=5), RCS (n=5), SWIPC with L-NNA (n=7), and RCS with L-NNA (n=5). All experiments were conducted in conscious pigs 1 to 2 weeks after recovery from surgery. SWIPC was induced by 2 cycles of 10-minute CAO and 10-minute coronary artery reperfusion (CAR). Lethal ischemia, ie, 60 minutes of CAO followed by 4 days of CAR, was performed for 24 hours after 2 cycles of ischemia. The RCS model was published previously.8,9 Briefly, ischemia was induced by constricting the left circumflex coronary artery to reduce the coronary blood flow by 35% to 45% from baseline. Coronary stenosis was maintained for 90 minutes and followed by reperfusion, repeated 6 times every 12 hours. After that, the same protocol of 60 minutes of CAO followed by 4 days of CAR was performed. Control pigs were subjected to the same CAO/CAR protocol. In both the SWIPC and RCS groups, a systemic infusion (intravenous) of L-NNA 35 mg/kg was given 1 hour before 60 minutes of CAO to inhibit NO synthase (NOS). All other procedures were the same as those for IPC and RCS. Separate pigs used for genomics and biochemical studies were subjected to SWIPC or RCS protocols but were not subjected to 60 minutes of lethal ischemia. Pigs subjected to SWIPC were euthanized 24 hours after IPC; pigs subjected to RCS were euthanized after 6 episodes of 90 minutes of coronary stenosis followed by reperfusion; and sham (control) pigs were euthanized without having undergone either IPC or lethal ischemia. All samples needed for protein and microarray analysis were taken from the subendocardium of the area at risk (AAR) in the different models and immediately frozen in liquid nitrogen. Thus, samples for biochemical and molecular analyses were taken at the same time as the lethal ischemia (60 minutes of CAO) was induced for the pigs used for studies of infarct size (IS) and hemodynamics, without the complicating factors of concurrent ischemia and infarction. The present study was approved by the Institutional Animal Care and Use Committee at the New Jersey Medical School.
Measurement of Cardiac Function
Measurements of global and regional myocardial function were recorded with a multiple-channel oscillograph. Aortic and left atrial pressures were measured with strain manometers that were calibrated with a mercury manometer connected to the fluid-filled catheters. The solid state LV pressure gauge was cross-calibrated with aortic and left atrial pressure measurements. LV dP/dt was obtained by electronically differentiating the LV pressure signal (Triton Technology Inc, San Diego, Calif). Anterior and posterior LV wall thicknesses were measured with an ultrasonic transit-time dimension gauge (Triton Technology Inc). LV end-diastolic dimensions for both regions were measured at the time that coincided with the beginning of the upstroke of the LV dP/dt. LV end-systolic dimensions were measured at minimum LV dP/dt. Systolic wall thickening was calculated as end-diastolic dimension minus end-systolic dimension. Mean coronary blood flow was measured with a Transonic flowmeter (Transonic Systems, Inc, Ithaca, NY).
Measurement of Infarct Size
Four days after CAR, the pigs were euthanized with an overdose of sodium pentobarbital, and the hearts were removed and placed on a dual-perfusion apparatus as described previously.10 Briefly, both the ascending aorta and the site of left circumflex CAO were cannulated and perfused retrogradely with Alcian blue (0.05%) and saline, respectively. After perfusion, the hearts were sliced into 6 to 8 rings, and both sides of the individual rings were photographed. The surface area of each ring was traced with computer-assisted Image-Pro software to measure the AAR and IS.
Microarray Data Analysis
cDNA synthesis (SuperScript, Invitrogen, Carlsbad, Calif) was performed from 10 µg of total RNA with a T7-oligo(24)dT primer. The DNA was transcribed into biotin-labeled RNA (bioarray RNA labeling kit, Enzo, New York, NY) and hybridized on a porcine genome array (Affymetrix, Santa Clara, Calif) that contained 23 937 probe sets targeting 20 201 genes of Sus scrofa. Human genes were used to annotate the porcine array.11 Gene Ontology (GO) annotations for genes were obtained from the National Center for Biotechnology Information gene database.12 The microarray data were normalized with the Robust Multichip Average method. We discarded probe sets that did not have signals above background in 75% of the arrays for all samples using the Affymetrix MAS5 present/ absent calls. The Significance Analysis of Microarrays (SAM) program was used to select probe sets that had differential signals between sample comparisons. A false-discovery rate of ≈5% and a median fold change >1.2 were used. GO entries were tested for bias for upregulation and downregulation of expression with the hypergeometric data distribution, as described previously.13 Each GO entry was given a significance score based on the probability values. When a GO entry has a more significant probability value for bias with upregulated genes, the significance score is −log10(probability value), or log10(probability value) otherwise.
Immunoblotting
Protein extracts were prepared from the LV of hearts with extraction buffer (20 mmol/L Tris-HCl [pH 7.4], 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1% Triton, 1 mmol/L sodium orthovanadate, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, and 5 µg/mL protease inhibitor cocktail). Protein concentration was determined by the Bradford method (Bio-Rad, Hercules, Calif). Proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and detected with specific antibodies. The blots were incubated with horseradish peroxidase–labeled secondary antibodies. Immunoreactive bands were detected with chemiluminescence (ECL, PerkinElmer Life Sciences, Waltham, Mass). The intensities of the resulting bands were quantified by Quantity One software on a GS-800 densitometer (Bio-Rad). Primary antibodies were used according to the manufacturer’s instructions. Western blotting for GAPDH was used to verify equal protein loading of the blots. NOS activity was measured with the Ultrasensitive Colorimetric NOS assay kit (Oxford Biomedical Research, Rochester Hills, Mich).
For PKCε translocation measurements, the cytosolic fraction was separated from the particulate fraction as described previously.14,15 Briefly, LV tissues were homogenized and centrifuged at 70 000g for 40 minutes at 4°C in lysis buffer containing 50 mmol/L Tris-HCl (pH 7.5), 5 mmol/L EDTA, 10 mmol/L EGTA, 1 mmol/L sodium orthovanadate, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, and 5 µg/mL protease inhibitor cocktail. The supernatant was taken as the cytosolic fraction. The pellet was resuspended and sonicated in lysis buffer mentioned above but also containing 1% Triton 100 and centrifuged under the same conditions. The supernatant was taken as the particulate fraction. The distribution of PKCε between the cytosolic and particulate fractions was measured by immunoblotting.
Statistical Analysis
Results are shown as the mean±SEM for the number of samples indicated in the Figure legends. ANOVA with Bonferroni post hoc correction was used for analysis of >2 groups. A value of P<0.05 was considered significant. The Student t test was used to select upregulated and downregulated genes (P<0.05 and fold change >1.2). The final gene set probability values were adjusted by Bonferroni correction for the microarray analysis.
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
Characterization of the Models
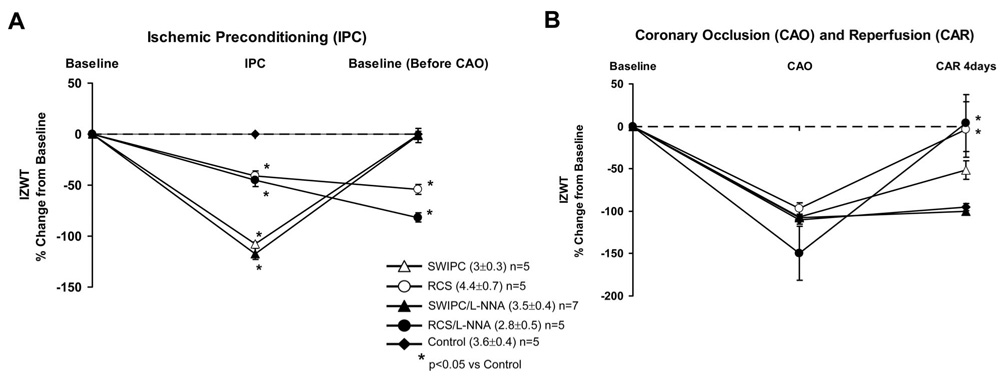
Hemodynamic data are presented in the Table. Hemodynamic data for the studies conducted after L-NNA administration are included in the Table in the Data Supplement. In the RCS model, during the first episode of coronary stenosis, coronary blood flow fell from 40±5 to 22±2 mL/min, and ischemic zone wall thickening (IZWT) fell by 40±7%, whereas anterior wall thickening, ie, the nonischemic zone, was not affected. The changes in IZWT are shown in Figure 1. Data after IPC are shown in Figure 1A, whereas the effects of lethal ischemia are shown in Figure 1B. IZWT remained depressed by 54±5% before the 60-minute period of lethal CAO, which reflects persistent myocardial dysfunction induced by chronic myocardial stunning, whereas coronary blood flow was 56±6 mL/min. Importantly, IZWT recovered at 4 days of CAR, after the 60-minute CAO (Figure 1B). In the SWIPC model, during the 2 episodes of complete CAO for 10 minutes to induce SWIPC, IZWT fell more than with RCS but recovered completely before the 60-minute CAO. SWIPC also showed some recovery of IZWT at 4 days after CAO/CAR. Again, there were no significant changes in nonischemic zone wall thickening.
Table.
Hemodynamic Data in Conscious Pigs
| % Change From Baseline |
|||||
|---|---|---|---|---|---|
| Baseline | IPC | Before CAO | CAO | CAR 4 d | |
| LV systolic pressure, mm Hg | |||||
| Control (n=5) | 115±8 | … | … | −11±5 | −8±6 |
| SWIPC (n=5) | 114±3 | −10±4 | −2±2 | −12±2 | −7±2 |
| RCS (n=5) | 121±5 | −1±2 | 1±4 | −8±4 | 1±5 |
| LV dP/dt max, mm Hg/s | |||||
| Control (n=5) | 2980±321 | … | … | −21±5 | −10±5 |
| SWIPC (n=5) | 3068±149 | −21±6 | −2±5 | −20±7 | 3±8 |
| RCS (n=5) | 3680±161 | −13±3 | −7±6 | −15±6 | −8±3 |
| Mean arterial pressure, mm Hg | |||||
| Control (n=5) | 96±5 | … | … | −9±6 | −2±5 |
| SWIPC (n=5) | 98±3 | −9±4 | −1±4 | −12±2 | −5±2 |
| RCS (n=5) | 98±5 | 1±3 | 5±4 | −5±4 | 4±5 |
| Heart rate, bpm | |||||
| Control (n=5) | 132±6 | … | … | −3±5 | −2±2 |
| SWIPC (n=5) | 132±5 | −9±4 | −4±4 | −9±2 | −3±4 |
| RCS (n=5) | 129±6 | 0±5 | 6±6 | −7±3 | 9±9 |
Figure 1.
A, Changes in IZWT during IPC and recovery from IPC (ie, baseline before lethal CAO) in control (sham), SWIPC, RCS, SWIPC/L-NNA, and RCS/L-NNA pigs. Absolute values for IZWT are shown in parentheses, and the number of pigs in each group is noted. Note that SWIPC resulted in a complete loss of function but returned to baseline after IPC, whereas RCS only partially reduced IZWT, but function remained depressed after RCS was completed. B, Changes in IZWT during the lethal 60-minute period of CAO followed by 4 days of reperfusion in all 5 groups. Note that 60 minutes of CAO resulted in a similar reduction in IZWT in all groups, but recovery was different. The greatest recovery of IZWT occurred in the RCS groups, consistent with reduced infarct size data shown in Figure 2.
RCS Confers Cardioprotection Equivalent to SWIPC
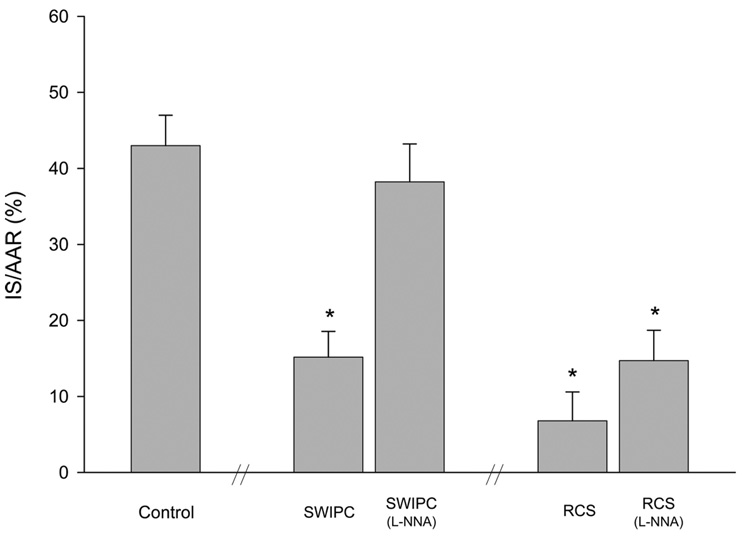
Lethal ischemia, induced by 60 minutes of CAO, resulted in an IS/AAR of 42±4% in control pigs. Whereas the AAR was comparable among groups, the IS/AAR was reduced significantly (P<0.05) in both the RCS model (6±3%) and the SWIPC model (16±3%; Figure 2).
Figure 2.
Reduction in infarct size conferred by SWIPC vs RCS. The Figure shows the reduction in IS/AAR in SWIPC and RCS compared with controls and shows that with addition of L-NNA, the cardioprotection was lost in SWIPC but not RCS. *P<0.05 vs control. n=5 per group except for SWIPC with L-NNA (n=7) and RCS with L-NNA (n=4). The dual perfusion of 1 pig heart in the RCS with L-NNA group was not adequate, and infarct size data could only be obtained in 4 animals in that group.
Molecular Differences Between RCS and SWIPC
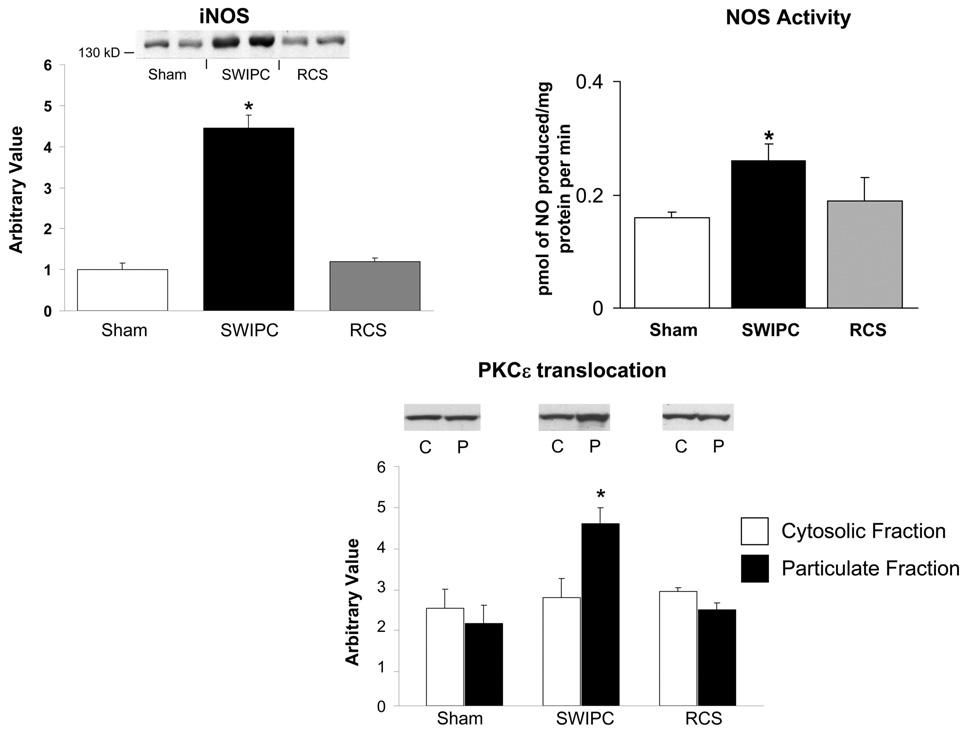
It is widely described that the cytoprotective mechanisms of IPC first require activation of PKCε on translocation to the plasma membrane, followed by a second window that includes increased expression of iNOS. We compared these molecular markers between the model of SWIPC and RCS studied here. Whereas translocation of PKCε and expression of iNOS were increased significantly in myocardium subjected to a protocol of SWIPC compared with sham, these markers were unaffected in myocardium subjected to RCS (Figure 3). Measurement of NOS activity (picomoles of NO produced per milligram of protein per minute) also confirmed the activation of NOS in SWIPC but not in RCS compared with sham (Figure 3).
Figure 3.
Mechanisms of SWIPC are not activated during RCS. Upregulation of iNOS, NOS activity, and the translocation of PKCε to the plasma membrane found during SWIPC are absent in the RCS model. *P<0.05 vs sham. Western blots show representative examples (n=5 for shams and n=6 for each SWIPC and RCS). C indicates cytosolic fraction; P, particulate fraction.
NOS Inhibition Prevents SWIPC but Not RCS
In agreement with previous reports,16 we confirmed that the cardioprotection conferred by SWIPC was abolished on NOS inhibition by L-NNA, ie, IS/AAR was no longer diminished after SWIPC (Figure 2). There was also a lack of recovery of IZWT (Figure 1B), consistent with the infarct data. However, the addition of L-NNA did not affect the protection conferred by RCS, in which IZWT recovered (Figure 1B) and IS/AAR remained significantly diminished (P<0.05; Figure 2). This confirms that the 2 methods of IPC protect the heart by different molecular mechanisms. The hemodynamics in the presence of L-NNA, as shown in Data Supplement Table I, were similar to those with SWIPC and RCS (Table), except that LV systolic pressure and arterial pressure were higher and heart rate was lower in the presence of L-NNA.
Gene Expression Differences in Cardioprotective Mechanisms
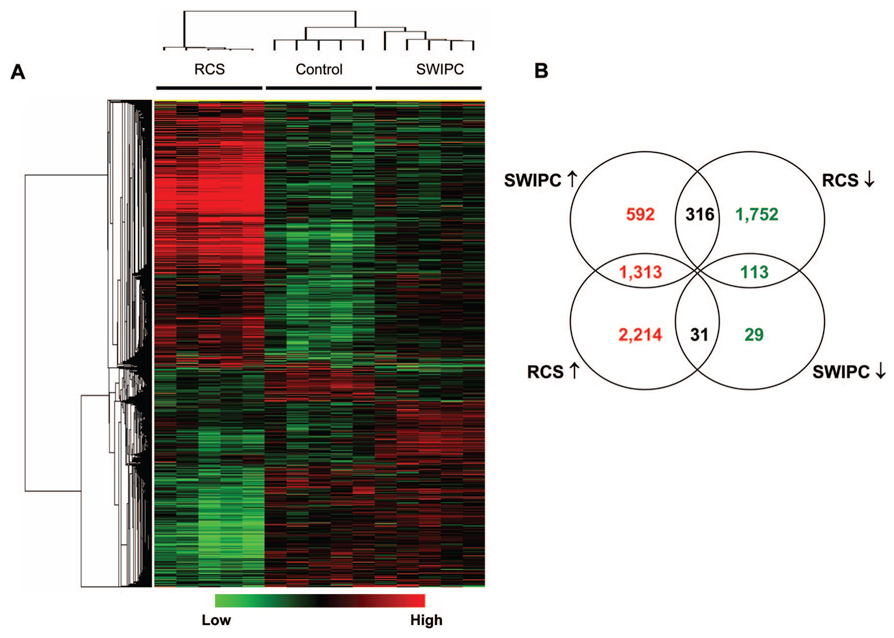
The results presented above provide evidence that the cyto-protective mechanisms activated by SWIPC and by RCS are qualitatively different. To further substantiate this conclusion, we performed a comprehensive analysis of gene expression using DNA microarrays. As shown in Figure 4, a large number of genes are differentially expressed in the 2 models. More genes were regulated in RCS (3558 upregulated and 2181 downregulated) than in SWIPC (2221 upregulated and 173 downregulated). A total of 1426 genes (1313 upregulated and 113 downregulated) were regulated in the 2 models with the same trend, ie, both upregulated or downregulated. A total of 621 genes (592 upregulated and 29 downregulated) were regulated in SWIPC but not in RCS, and 3966 genes (2214 upregulated and 1752 downregulated) were regulated in RCS but not in SWIPC. Only 347 genes showed reciprocal regulation between both models.
Figure 4.
Differentially expressed genes from 2 models. A, Probe sets for genes that are significantly regulated (fold change >1.2 and q-value <0.05 by significance analysis of microarrays) in either SWIPC or RCS were selected, clustered, and shown in a heat map. The log2-based values were median-centered in each row and are represented according to the color scale shown at the bottom. B, Venn diagram that summarizes the similarities and differences between the 2 models using significant genes in Figure 4A. When multiple probe sets were present for a gene, the most significant one, either upregulated or downregulated, was selected. n=5 per group.
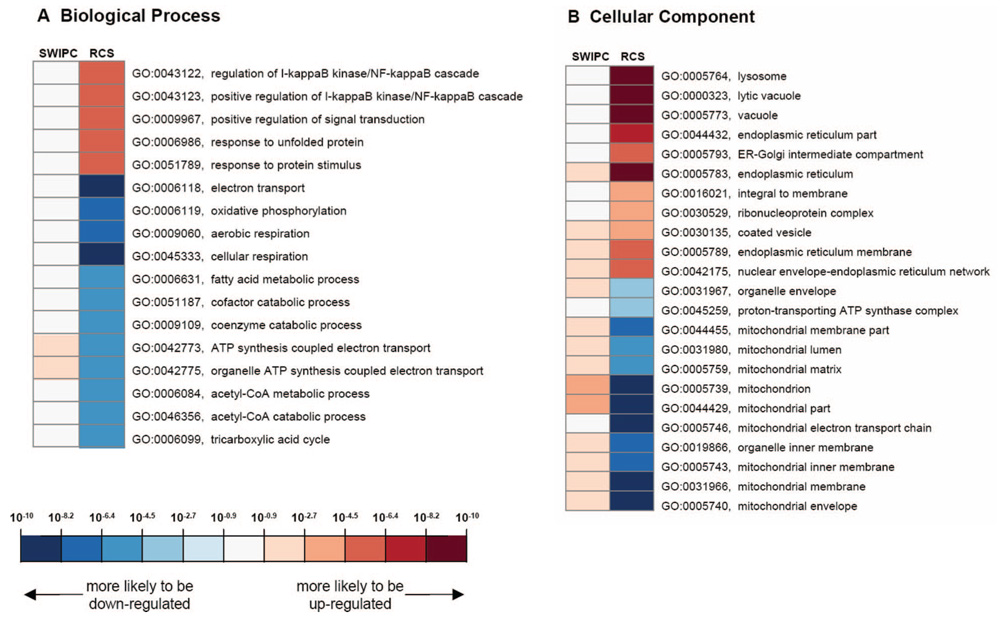
To understand the gene pathways that are regulated in the 2 models, we analyzed regulated genes by their GO annotations, including the biological process (BP), molecular function (MF), and cellular component (CC) categories. Representative highly significant GO entries for BP and CC are shown in Figure 5. We found that SWIPC and RCS models were conspicuously different with respect to regulation of gene pathways. Genes related to mitochondrial energy metabolism (several GO entries in Figure 5A and 5B) tend to be downregulated in RCS, whereas genes involved in cardiac protection, including autophagy (“lysosome” and “lytic vacuole” in Figure 5B) and endoplasmic reticulum (ER) stress response (several entries related to ER in Figure 5B), tend to be upregulated in RCS.
Figure 5.
Significant GO entries were selected by the hypergeometric test (<0.05 after Bonferroni correction) for SWIPC and RCS models and are shown in a heat map with color representing the significance score (see Methods for its calculation). As shown in the color scale at the bottom, GO entries with positive significance scores (greater significant probability values for biased representation of upregulated genes) are shown in red, and those with negative scores (greater significant probability values for biased representation of downregulated genes) are shown in blue. To avoid redundancy in the graph, GO entries with >100 child entries are not shown. NF indicates nuclear factor; CoA, coenzyme A.
Verification of Cardioprotective Mechanisms Derived From Microarray Analysis
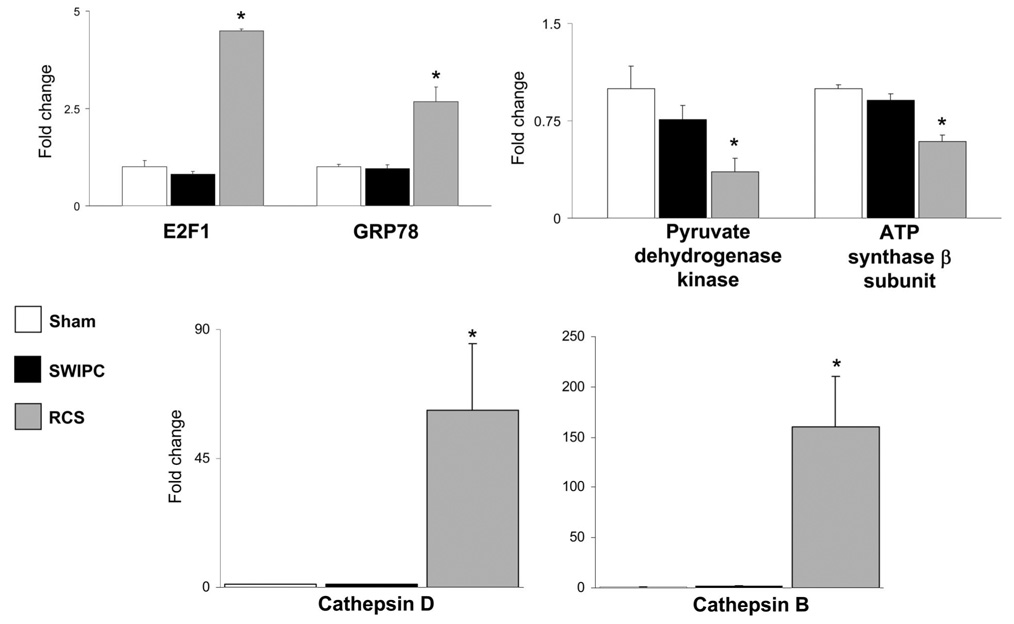
We verified the genomic differences described above at the protein level by measuring representative examples of the regulated gene sets participating in cardioprotective mechanisms, such as the mediators of autophagy, cathepsins B and D, and the ER chaperone protein GRP78. The expression of these proteins was compared by Western blotting in LV samples from sham, SWIPC, and RCS pigs. There was no significant change in the expression of cathepsins B and D or GRP78 in the model of SWIPC compared with sham, whereas these proteins were increased significantly in the model of RCS (Figure 6). Reciprocally, the microarray data show a global downregulation of mitochondrial oxidative pathways only in the RCS model (Figure 5). Accordingly, pyruvate dehydrogenase kinase and ATP synthase-β were downregulated significantly in the model of RCS but not in SWIPC (Figure 6). These results confirm that the transcriptional regulation found by microarrays is accompanied by regulation of the corresponding proteins.
Figure 6.
Regulation of proteins involved in cardioprotection specific to RCS. There was no significant change (data expressed as fold change from sham) in the expression of cathepsin D, E2F1, and GRP78 in the model of SWIPC compared with sham, but they were increased significantly by 20-, 4.5-, 4.0-, and 4.2-fold, respectively, in the RCS model (P>0.05 vs SWIPC). Cathepsin B was increased by 100-fold in RCS (P<0.01 vs SWIPC). Reciprocally, pyruvate dehydrogenase kinase and ATP synthase-β were downregulated in RCS but not in SWIPC. n=6 per group, except for cathepsins in RCS, n=7. *P<0.05 vs SWIPC.
The upregulation of autophagy and ER stress genes in RCS suggests a potential protection mechanism that involves cell survival. In fact, genes associated with cell cycle–related GO entries and the cell cycle regulator E2F1 were significantly upregulated in RCS but not in SWIPC. Thus, upregulation of genes in autophagy, ER stress, cell cycle, and cell survival, together with downregulation of genes in mitochondrial function, defines the cardiac protection mechanism by the RCS model and distinguishes it from the SWIPC model.
Discussion
The major finding of the present investigation is that RCS induces cardioprotection that is quantitatively at least equal to that of the more traditionally used IPC protocols (SWIPC), but the mechanisms mediating this protection are radically different in the RCS model. Most importantly, it is widely accepted that PKC and NO are cornerstones of the cardioprotection induced by SWIPC2–7 and that pretreatment with an NOS inhibitor can abolish the IPC. These cardinal observations were confirmed in the model of SWIPC used in the present investigation. In contrast, the protection afforded by RCS was not accompanied by upregulation of either PKC or iNOS and was not diminished by pretreatment with the NOS inhibitor. Once we recognized that the mechanisms mediating IPC were so disparate to those induced by RCS, we conducted microarray analysis to determine the extent of differences in the genomic signature of SWIPC compared with RCS, with the hypothesis that novel mechanisms mediating RCS would be discovered with this approach.
Indeed, a possible explanation for these marked mechanistic differences may come from the comparison of the genomic profile between the models used in the present study. The present results indicate that the model of RCS regulates specific categories of genes that are not affected by SWIPC. This model-specific genomic regulation includes a downregulation of genes involved in mitochondrial energy metabolism and an upregulation of genes that participate in 3 pathways of cardiac cytoprotection, ie, autophagy, ER stress response, and cell survival (Figure 6).
Multiple genes involved in mitochondrial oxidative function are downregulated by RCS, which is in agreement with a recent study conducted by 2D gel electrophoresis showing a similar downregulation of mitochondrial proteins in a model of long-term (3 months) coronary artery stenosis.17 Several studies have illustrated the importance of the production of reactive oxygen species from mitochondria to explain the mechanisms of IPC.18–20 It is therefore possible that a shutdown of mitochondrial function in repetitive ischemia will elicit the activation of reactive oxygen species–independent and totally different survival mechanisms. These alternative mechanisms include autophagy, ER stress response, and cell survival. At this point, we do not have definitive evidence of the extent of cardioprotection induced by these mechanisms. Future work will be required to elucidate the relative importance of these pathways, including mitochondrial mechanisms; however, this will be complicated by the possibility that several mechanisms mediating protection after RCS could be redundant. This would not be unexpected in view of the large number of genes (5739) that were found to be regulated in the heart in the RCS model.
Autophagy is an intracellular process of degradation of damaged organelles and denatured cytoplasmic proteins that reduces cellular stress and promotes cell survival.21 We recently showed that activation of the autophagy pathway of protein degradation is a survival mechanism in the chronically ischemic myocardium characterized by autophagosomes in myocytes, as seen with electron microscopy, and by upregulation of proteins involved in autophagy.9 Although autophagy can disrupt myocytes, the salvaged amino acids can be used to build new proteins, whereas necrosis and apoptosis result in cell death without regeneration. The lysosomal cathepsins B and D are central to the mechanisms of autophagy, because their deletion leads to an inhibition of autophagy and accumulation of denatured proteins.22 The present results confirm a major upregulation of both enzymes in the model of RCS, but this was not observed in SWIPC.
The ER stress response is a mechanism that improves the quality of protein translation by limiting the production and accelerating the degradation of denatured peptides by the proteasome.23 The ER is crucial for protein synthesis and secretion, but a high rate of translation automatically includes a large proportion of misfolded and denatured proteins that must be destroyed. ER-specific chaperones, such as GRP78, prevent the accumulation of unfolded proteins in the ER by promoting their translocation to the ubiquitin-proteasome system of protein degradation, a process known as ER-associated degradation.23 If the ER-associated degradation is saturated, there will be a rapid accumulation of unfolded proteins in the ER, which triggers the “unfolded protein response” that results in the death of the cell by apoptosis.23 In that respect, autophagy and ER stress response are complementary in preventing the accumulation of denatured proteins that would automatically increase cellular stress.
The mechanistic differences between SWIPC and RCS described in the present study have profound clinical implications. As discussed above, the model of RCS resembles more closely the clinical condition of patients with ventricular dysfunction that results from sequential episodes of stress-induced stable angina.24 Furthermore, although preconditioning has been the subject of intense investigation in various animal models, its relevance in the clinical setting is less established, which can be attributed in part to the fact that differences in mechanisms mediating cardioprotection are radically different in the presence of chronic, repetitive ischemia than after a traditional IPC stimulus applied to virgin myocardium.
One other difference between traditional IPC and the RCS-induced ischemic protection must be mentioned, ie, preconditioning generally but not always25–27 follows episodes of completed CAO and reperfusion and not coronary stenosis. Accordingly, we examined 3 additional pigs with a protocol of two 10-minute periods of CAO followed by CAR, which was repeated every 12 hours 6 times. IS/AAR was 12±3% (data not shown), similar to that observed with RCS (6±3%) or SWIPC (16±4%). After L-NNA, in 3 additional pigs with this protocol, ischemic protection was not abolished, ie, IS/AAR was 15±2%, similar to that observed with RCS and L-NNA (Figure 2). Thus, although NOS is critical to the second window of IPC, it is not involved in mediating the IPC elicited by repetitive episodes of either coronary stenosis or complete coronary occlusion. However, this does not completely rule out a role for NO, because it was recently shown that NO can be produced in the ischemic heart independently from NOS activity.28
Another mechanism of IPC has been described with coronary microembolization over a 6-hour period.29 Microinfarction and inflammation occur, which results in IPC mediated by tumor necrosis factor-α. Even though this occurs temporally before SWIPC (6 hours versus 24 hours), it was termed a “third window” of IPC.25 The RCS model also is characterized by sparse, focal lesions of necrosis, primarily in the subendocardium.8 Therefore, it is possible that mechanisms related to inflammation may be involved in the cardioprotection, but as noted above, the microarray analysis identified 5739 genes regulated in the RCS model. Accordingly, it is not likely that 1 mechanism can be responsible for cardioprotection with chronic, repetitive episodes of ischemia.
In conclusion, RCS induces powerful protection against lethal myocardial ischemia, equivalent to that induced by traditional IPC but which acts through mechanisms radically different from those observed during preconditioning. These data demonstrate the existence of a novel “third window” of cardioprotection, which is likely involved in the protection inherent to chronic and repetitive ischemia found in patients with coronary artery disease.
CLINICAL PERSPECTIVE
Ischemic preconditioning (IPC), discovered 20 years ago, is the most powerful intervention known to protect myocardium, and yet it has proved difficult to translate the knowledge obtained into clinical therapy. The vast majority of experimental studies in IPC have used a brief episode of the IPC stimulus on a background of a normal heart with normal coronary arteries, ie, virgin ischemia. Our hypothesis is that the molecular mechanisms that mediate IPC are radically different in the setting of chronic ischemia, more akin to the situation in patients with coronary artery disease. To test this, conscious, chronically instrumented pigs were subjected to either repetitive coronary stenosis (RCS) or a traditional protocol of second-window IPC (SWIPC). Lethal ischemia, applied after IPC, resulted in similar reductions in infarct size/area at risk for animals in the RCS and SWIPC protocols. Two molecular signatures of SWIPC, the increased expression of the inducible isoform of NO synthase and the translocation of protein kinase Cε to the plasma membrane, were observed with SWIPC but not with RCS. Microarray analysis revealed a qualitatively different genomic profile of cardioprotection between IPC induced by RCS and that induced by SWIPC. The number of genes significantly regulated was greater in RCS than in SWIPC. Therefore, RCS induces cardioprotection against lethal myocardial ischemia that is at least as powerful as traditional IPC but is mediated through radically different mechanisms.
Supplementary Material
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/cgi/content/full/CIRCULATIONAHA.108.788240/DC1.
Acknowledgments
We appreciate the helpful editorial assistance provided by Lauren Danridge.
Sources of Funding
This work was supported in part by National Institutes of Health grants AG014121, HL033107, HL059139, HL069752, AG027211, AG023137, HL069020, and AG023567.
Footnotes
Disclosures
None.
References
- 1.Murry CE, Jennings RB, Reimer KA. Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation. 1986;74:1124–1136. doi: 10.1161/01.cir.74.5.1124. [DOI] [PubMed] [Google Scholar]
- 2.Bolli R, Manchikalapudi S, Tang XL, Takano H, Qiu Y, Guo Y, Zhang Q, Jadoon AK. The protective effect of late preconditioning against myocardial stunning in conscious rabbits is mediated by nitric oxide synthase: evidence that nitric oxide acts both as a trigger and as a mediator of the late phase of ischemic preconditioning. Circ Res. 1997;81:1094–1107. doi: 10.1161/01.res.81.6.1094. [DOI] [PubMed] [Google Scholar]
- 3.Guo Y, Jones WK, Xuan YT, Tang XL, Bao W, Wu WJ, Han H, Laubach VE, Ping P, Yang Z, Qiu Y, Bolli R. The late phase of ischemic preconditioning is abrogated by targeted disruption of the inducible NO synthase gene. Proc Natl Acad Sci U S A. 1999;96:11507–11512. doi: 10.1073/pnas.96.20.11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Ytrehus K, Downey JM. Evidence that translocation of protein kinase C is a key event during ischemic preconditioning of rabbit myocardium. J Mol Cell Cardiol. 1994;26:661–668. doi: 10.1006/jmcc.1994.1078. [DOI] [PubMed] [Google Scholar]
- 5.Qiu Y, Ping P, Tang XL, Manchikalapudi S, Rizvi A, Zhang J, Takano H, Wu WJ, Teschner S, Bolli R. Direct evidence that protein kinase C plays an essential role in the development of late preconditioning against myocardial stunning in conscious rabbits and that epsilon is the isoform involved. J Clin Invest. 1998;101:2182–2198. doi: 10.1172/JCI1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saurin AT, Pennington DJ, Raat NJ, Latchman DS, Owen MJ, Marber MS. Targeted disruption of the protein kinase C epsilon gene abolishes the infarct size reduction that follows ischaemic preconditioning of isolated buffer-perfused mouse hearts. Cardiovasc Res. 2002;55:672–680. doi: 10.1016/s0008-6363(02)00325-5. [DOI] [PubMed] [Google Scholar]
- 7.Takano H, Manchikalapudi S, Tang XL, Qiu Y, Rizvi A, Jadoon AK, Zhang Q, Bolli R. Nitric oxide synthase is the mediator of late preconditioning against myocardial infarction in conscious rabbits. Circulation. 1998;98:441–449. doi: 10.1161/01.cir.98.5.441. [DOI] [PubMed] [Google Scholar]
- 8.Kim SJ, Peppas A, Hong SK, Yang G, Huang Y, Diaz G, Sadoshima J, Vatner DE, Vatner SF. Persistent stunning induces myocardial hibernation and protection: flow/function and metabolic mechanisms. Circ Res. 2003;92:1233–1239. doi: 10.1161/01.RES.0000076892.18394.B6. [DOI] [PubMed] [Google Scholar]
- 9.Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci U S A. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen YT, Knight DR, Vatner SF, Randall WC, Thomas JX., Jr Responses to coronary artery occlusion in conscious dogs with selective cardiac denervation. Am J Physiol. 1988;255:H525–H533. doi: 10.1152/ajpheart.1988.255.3.H525. [DOI] [PubMed] [Google Scholar]
- 11.Tsai S, Cassady JP, Freking BA, Nonneman DJ, Rohrer GA, Piedrahita JA. Annotation of the Affymetrix porcine genome microarray. Anim Genet. 2006;37:423–424. doi: 10.1111/j.1365-2052.2006.01460.x. [DOI] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology: the Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu H, Tian B, Resuello RG, Natividad FF, Peppas A, Shen YT, Vatner DE, Vatner SF, Depre C. Sex-specific regulation of gene expression in the aging monkey aorta. Physiol Genomics. 2007;29:169–180. doi: 10.1152/physiolgenomics.00229.2006. [DOI] [PubMed] [Google Scholar]
- 14.Ping P, Zhang J, Qiu Y, Tang XL, Manchikalapudi S, Cao X, Bolli R. Ischemic preconditioning induces selective translocation of protein kinase C isoforms epsilon and eta in the heart of conscious rabbits without subcellular redistribution of total protein kinase C activity. Circ Res. 1997;81:404–414. doi: 10.1161/01.res.81.3.404. [DOI] [PubMed] [Google Scholar]
- 15.Wolfrum S, Schneider K, Heidbreder M, Nienstedt J, Dominiak P, Den-dorfer A. Remote preconditioning protects the heart by activating myocardial PKCepsilon-isoform. Cardiovasc Res. 2002;55:583–589. doi: 10.1016/s0008-6363(02)00408-x. [DOI] [PubMed] [Google Scholar]
- 16.Bolli R. The late phase of preconditioning. Circ Res. 2000;87:972–983. doi: 10.1161/01.res.87.11.972. [DOI] [PubMed] [Google Scholar]
- 17.Page B, Young R, Iyer V, Suzuki G, Lis M, Korotchkina L, Patel MS, Blumenthal KM, Fallavollita JA, Canty JM., Jr Persistent regional down-regulation in mitochondrial enzymes and upregulation of stress proteins in swine with chronic hibernating myocardium. Circ Res. 2008;102:103–112. doi: 10.1161/CIRCRESAHA.107.155895. [DOI] [PubMed] [Google Scholar]
- 18.Kaiserova K, Srivastava S, Hoetker JD, Awe SO, Tang XL, Cai J, Bhatnagar A. Redox activation of aldose reductase in the ischemic heart. J Biol Chem. 2006;281:15110–15120. doi: 10.1074/jbc.M600837200. [DOI] [PubMed] [Google Scholar]
- 19.Oldenburg O, Qin Q, Krieg T, Yang XM, Philipp S, Critz SD, Cohen MV, Downey JM. Bradykinin induces mitochondrial ROS generation via NO, cGMP, PKG, and mitoKATP channel opening and leads to cardioprotection. Am J Physiol Heart Circ Physiol. 2004;286:H468–H476. doi: 10.1152/ajpheart.00360.2003. [DOI] [PubMed] [Google Scholar]
- 20.Tang XL, Takano H, Rizvi A, Turrens JF, Qiu Y, Wu WJ, Zhang Q, Bolli R. Oxidant species trigger late preconditioning against myocardial stunning in conscious rabbits. Am J Physiol Heart Circ Physiol. 2002;282:H281–H291. doi: 10.1152/ajpheart.2002.282.1.H281. [DOI] [PubMed] [Google Scholar]
- 21.Yan L, Sadoshima J, Vatner DE, Vatner SF. Autophagy: a novel protective mechanism in chronic ischemia. Cell Cycle. 2006;5:1175–1177. doi: 10.4161/cc.5.11.2787. [DOI] [PubMed] [Google Scholar]
- 22.Qin ZH, Wang Y, Kegel KB, Kazantsev A, Apostol BL, Thompson LM, Yoder J, Aronin N, DiFiglia M. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum Mol Genet. 2003;12:3231–3244. doi: 10.1093/hmg/ddg346. [DOI] [PubMed] [Google Scholar]
- 23.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Depre C, Vatner SF. Cardioprotection in stunned and hibernating myocardium. Heart Fail Rev. 2007;12:307–317. doi: 10.1007/s10741-007-9040-3. [DOI] [PubMed] [Google Scholar]
- 25.Heusch P, Skyschally A, Leineweber K, Haude M, Erbel R, Heusch G. The interaction of coronary microembolization and ischemic preconditioning: a third window of cardioprotection through TNF-alpha. Arch Med Sci. 2007;3:83–92. [Google Scholar]
- 26.Domenech R, Macho P, Schwarze H, Sanchez G. Exercise induces early and late myocardial preconditioning in dogs. Cardiovasc Res. 2002;55:561–566. doi: 10.1016/s0008-6363(02)00334-6. [DOI] [PubMed] [Google Scholar]
- 27.Domenech RJ. Preconditioning: a new concept about the benefit of exercise. Circulation. 2006;113:e1–e3. doi: 10.1161/CIRCULATIONAHA.105.569863. [DOI] [PubMed] [Google Scholar]
- 28.Martin C, Schulz R, Post H, Boengler K, Kelm M, Kleinbongard P, Gres P, Skyschally A, Konietzka I, Heusch G. Microdialysis-based analysis of interstitial NO in situ: NO synthase-independent NO formation during myocardial ischemia. Cardiovasc Res. 2007;74:46–55. doi: 10.1016/j.cardiores.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 29.Skyschally A, Gres P, Hoffmann S, Haude M, Erbel R, Schulz R, Heusch G. Bidirectional role of tumor necrosis factor-alpha in coronary microembolization: progressive contractile dysfunction versus delayed protection against infarction. Circ Res. 2007;100:140–146. doi: 10.1161/01.RES.0000255031.15793.86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/cgi/content/full/CIRCULATIONAHA.108.788240/DC1.