Abstract
Context:
There is evidence of linkage to a schizophrenia susceptibility locus on chromosome 8p21-22 found by several family linkage studies.
Objectives:
To fine map and identify a susceptibility gene for schizophrenia on chromosome 8p22 and to investigate the effect of this genetic susceptibility on an endophenotype of abnormal brain structure using magnetic resonance imaging.
Design:
Fine mapping and identification of a chromosome 8p22 susceptibility gene was carried out by finding linkage disequilibrium between genetic markers and schizophrenia in multiply affected families, a case-control sample, and a trio sample. Variation in brain morphology associated with pericentriolar material 1 (PCM1) alleles was examined using voxel-based morphometry and statistical parametric mapping with magnetic resonance imaging.
Setting and Patients:
A family sample of 13 large families multiply affected with schizophrenia, 2 schizophrenia case-control samples from the United Kingdom and Scotland, and a sample of schizophrenic trios from the United States containing parents and 1 affected child with schizophrenia.
Main Outcome Measures:
Tests of transmission disequilibrium between PCM1 locus polymorphisms and schizophrenia using a family sample and tests of allelic association in case-control and trio samples. Voxel-based morphometry using statistical parametric mapping.
Results:
The family and trio samples both showed significant transmission disequilibrium between marker D85261 in the PCM1 gene locus and schizophrenia. The case-control sample from the United Kingdom also found significant allelic association between PCM1 gene markers and schizophrenia. Voxel-based morphometry of cases who had inherited a PCM1 genetic susceptibility showed a significant relative reduction in the volume of orbitofrontal cortex gray matter in comparison with patients with non–PCM1–associated schizophrenia, who, by contrast, showed gray matter volume reduction in the temporal pole, hippocampus, and inferior temporal cortex.
Conclusions:
The PCM1 gene is implicated in susceptibility to schizophrenia and is associated with orbitofrontal gray matter volumetric deficits.
The evidence of a major role for genetic susceptibility in the etiology of schizophrenia has been shown by family, twin, adoption, and genetic linkage studies. Rather than the family environment, specific, unique environmental factors, which include biological environment and stochastic effects that are not shared by siblings, have been implicated.1 Two types of investigation have dominated research into schizophrenia. The first type, using structural brain imaging studies, has consistently shown volumetric abnormalities in the brains of patients with schizophrenia. The other involves the systematic mapping of chromosomal regions in which genes that increase susceptibility to schizophrenia are present. A key finding has been the presence of locus heterogeneity with different genetic susceptibilities being involved in the etiology of schizophrenia in different multiply affected families.
Genetic linkage analyses to a chromosome 8p21-22 schizophrenia susceptibility locus (SCZD6, Online Mendelian Inheritance in Man [OMIM] 603013), under the assumption of locus heterogeneity or admixture, confirm that this region of chromosome 8 harbors a susceptibility gene for schizophrenia. Three independent investigators2-4 report lod scores above 3.00 and a fourth5 confirms a lod score above 2.00. Once linkage has generally implicated a region of a chromosome, it is necessary to fine map exactly which gene is involved by tests of transmission disequilibrium and allelic association with schizophrenia to detect underlying linkage disequilibrium between marker alleles and disease alleles.
In an earlier attempt to fine map a specific susceptibility gene on 8p22, we carried out tests of allelic association in a smaller case-control sample with 15 markers that flanked the PCM1 gene without finding any evidence of association.6 We then carried out tests of transmission disequilibrium of markers genotyped in our family study and followed up these results in 2 case-control samples and then in a collection of trios consisting of both parents and 1 affected offspring. Once allelic association due to linkage disequilibrium between markers and schizophrenia is detected, it is possible to differentiate genetic subgroups of schizophrenia and to examine clinical or endophenotypic characteristics of genotypically defined groups. We chose the endophenotype of abnormal brain morphology to study in relation to genotype because it seemed a priori the most likely variable to be associated with underlying heterogeneous genetic effects.
The evidence that there are volumetric deficits in the brains of subjects with schizophrenia has been replicated many times both in first-onset illness and in chronically affected subjects.7-11 Consistency is generally reported in the regional pattern of cortical and subcortical morphometric abnormalities affecting gray matter density and volume.9,10 The diagnosis of schizophrenia encompasses symptom clusters putatively associated with dysfunction of distinct neuroanatomical substrates.12 Methodological advances now enable unbiased automated analysis of structural magnetic resonance imaging (MRI) brain scans on a voxel-wide basis to quantify differences in brain morphometry and tissue composition at a level of detail that cannot be achieved by macroscopic examination.13-19 In this study, we describe an application of voxel-based morphometry (VBM) to test for differences in regional brain volume in patients with 8p22 PCM1–associated schizophrenia. A chromosome 8 subgroup was selected on the basis of having inherited specific marker alleles at the PCM1 gene locus and compared with patients with schizophrenia without these alleles and with healthy matched control subjects.
METHODS
PATIENT AND CONTROL SAMPLES
The clinical sampling, diagnosis, and genotyping of the family sample for the testing of transmission disequilibrium between alleles at the D8S261 locus and schizophrenia has been described previously.4 Briefly, it consisted of 13 large, affected pedigrees containing cases diagnosed using Research Diagnostic Criteria (RDC). Pedigrees were selected on the basis of appearing to demonstrate a single source of schizophrenia or DSM-III-R schizotypal illness with unilineal transmission and no cases of bipolar affective disorder. A pedigree was considered unilineal not just on the basis of the interview diagnoses with the Schedule for Affective Disorders and Schizophrenia–Lifetime Version (SADS-L) but also with all the information available from past records. Two affection classes were used for the extended transmission disequilibrium test (ETDT) and linkage analyses: core schizophrenia consists of schizophrenia, unspecified functional psychosis, and schizoaffective psychosis; schizophrenia spectrum consists additionally of schizoid and schizotypal personality disorder according to DSM-III-R criteria. Of the 182 interviewed individuals, 56 fell into the core schizophrenia category and an additional 12 fell into the spectrum category.
The sampling of the UK case-control sample has recently been described in a study showing a genetic effect on schizophrenia from the region of the epsin 4 gene on chromosome 5.20 We recruited a sample of unrelated volunteers with schizophrenia and unrelated “supernormal” controls from London and South England. This type of control, in which mental illness in the control subjects and their first-degree relatives is grounds for exclusion, will maximize the chance of finding a genetic difference between a schizophrenic group and a control group. All subjects in the case-control samples and the subsamples used for MRI scanning were included only if both parents were of white English, Irish, Welsh, or Scottish descent and if 3 out of 4 grandparents were of the same descent. One grandparent was allowed to be of Caucasian European origin but not of Jewish or non–European Union ancestry based on the European Union countries before the recent enlargement. Approval was obtained from the UK National Health Service multicenter and local research ethics boards, and all subjects signed an approved consent form after reading an information sheet.
All 450 schizophrenic cases were first selected for having a clinical diagnosis of schizophrenia made by a psychiatrist recorded in National Health Service medical case notes according to the International Classification of Diseases, 10th Revision (ICD-1 0), which is the same as the DSM-IV diagnosis of schizophrenia. Subjects with clear organic or symptomatic schizophreniform psychoses were excluded. All subjects were interviewed and data were collected with the SADS-L interview schedule.21 Accuracy of data entries into the SADS-L was ensured by using medical case note information and all other available records. All cases were diagnosed as having schizophrenia according to the probable level of the RDC. The supernormal control subjects were also interviewed with the initial clinical screening questions of the SADS-L and selected on the basis of not having a family history of schizophrenia, alcoholism, or bipolar disorder and for having no past or present personal history of any RDC-defined mental disorder.
The US trio sample consisted of 100 unrelated patients meeting DSM-III-R criteria for schizophrenia or schizoaffective disorder as determined by structured diagnostic interview (Structured Clinical Interview for DSM-III-R [SCID]) conducted by trained raters with demonstrated interrater reliability. The research subjects were recruited from the Experimental Therapeutics branch of the National Institute of Mental Health and the Maryland Psychiatric Research Center of the University of Maryland School of Medicine, Baltimore. All subjects gave written informed consent to participate in the study under a protocol approved by the institutional review boards of the National Institute of Mental Health and the University of Maryland School of Medicine. After we obtained informed consent, the parents of each subject were also recruited for genetic testing.
The Scottish sample consisted of 200 unrelated cases of schizophrenia and 200 unrelated controls. Ethical approval was granted by the University of Edinburgh and the Royal Edinburgh Hospital, and all subjects gave informed written consent. These cases were diagnosed using the same clinical assessment (SADS-L) and diagnostic system (RDC) as those of the London sample. Accuracy of data entries into the SADS-L was ensured by using medical case note information and all other available records. The information on all cases was reviewed by a second psychiatrist who confirmed the RDC diagnoses. All cases were diagnosed as having schizophrenia according to the probable level of the RDC. The Scottish normal control subjects were also interviewed with the SADS-L and selected on the basis of not having a family history of schizophrenia, alcoholism, or bipolar disorder and for having no past or present personal history of any RDC-defined mental disorder. Ancestry was checked and all subjects were of white English, Irish, Scottish, or Welsh origin. The subgroups of research subjects with schizophrenia and controls employed for the MRI study were all selected only from London-based schizophrenic or control samples, and the imaging research was carried out with approval from the National Health Service multicenter research ethics committee, the National Hospital for Neurology and Neurosurgery and Institute of Neurology Joint Research Ethics Committee (University College London Hospitals National Health Service Trust), and the University College London Hospitals ethics committee.
GENETIC METHODS
Novel CA dinucleotide repeat polymorphisms were identified by screening genomic DNA close to the marker D8S261. Primers to amplify these repeats were designed using the Primer3 software program.22 The 3 (CA) n repeat markers were D8S261, D8S2615, and D8S2616. Their repeat fragment sizes, respective allele numbers, and heterozygosities are listed at http://www.gdb.org (the Genome Database). Genotyping methods for the single nucleotide polymorphism markers rs916550, rs4 12750, rs454755, rs208753, rs3780 103, rs699 1775, rs445422, rs412750, rs13276297, rs3214087, and rs370429 are at Ensembl, http://www.ensembl.org; the University of California, Santa Cruz, genome browser, http://www.genome.ucsc.edu/; and the Single Nucleotide Polymorphism database at the National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/projects/SNP/. Polymerase chain reaction conditions for amplification were as previously described.6 The familial test of association was computed with the ETDT.23 The CLUMP test of allelic association24 was used for the case-control samples. All P values reported (including the ETDT test of the 100 US trios) are empirically derived using a Monte Carlo simulation approach. Subtests of the CLUMP program were the following: T1, Pearson x2 statistic of the raw contingency table; T2, the x2 statistic of a table with rare alleles grouped together to prevent small expected cell counts; T3, the largest of the x2 statistics of 2 × 2 tables each of which compares 1 allele against the rest grouped together; T4, the largest of the x2 statistics of all possible 2 × 2 tables comparing any combination of alleles against the rest. Tests of haplotype association were carried out with GENECOUNTING/LDPAIRS and confirmed using an empirical permutation test of significance.25,26 The extent of linkage disequilibrium between markers was calculated with estimated haplotypes as incorporated into GENECOUNTING/LDPAIRS25-27 (Figure 1).
Figure 1.

Tests of pairwise linkage disequilibrium between markers within the PCM1gene.
VOXEL-BASED MORPHOMETRY
There were 2 patient groups. One group with schizophrenia (SZ8) was selected on the basis that they had inherited marker alleles from any 2 markers that were associated with schizophrenia. This group comprised 14 subjects (mean±SD age, 40.9±8.4 years; 6 male and 8 female patients). A second patient group also had schizophrenia (SZ0) but had not inherited any of the alleles associated with chromosome 8 schizophrenia (n= 13; mean±SD age, 42.5 ±8.4 years; 10 male and 2 female patients). All SZ8 and SZ0 schizophrenic research subjects had diagnoses of chronic or subchronic schizophrenia and at the time of scanning were all being treated with antipsychotic drugs. One of 14 SZ8 cases was recorded as being unresponsive to treatment. Two of 13 SZ0 cases were recorded as being unresponsive to treatment. All other SZ8 and SZ0 cases were partially responsive to treatment. None had good prognosis schizophrenia, and all were unemployed.
The mean± SD age at onset for the SZ8 PCM1-associated group was 19.1±5.7 years; the mean± SD age at onset for the SZ0 group with non–PCM1 -associated schizophrenia was 18.8±3.1 years. Healthy control subjects, who were screened to exclude medication use and psychological, neurological, or systemic illness, were individually matched with patients with schizophrenia by age, sex, social class, and ethnicity. These research subjects were not genotyped and were not part of the normal control groups used in the case-control sample. These included 14 normal controls matched to patients with chromosome 8–associated schizophrenia (CON8) (mean±SD age, 39.8± 10.0 years; 6 men and 8 women) and 13 normal controls matched to the patients with non–chromosome 8 schizophrenia (CON0) (mean±SD age, 42.7±11.7 years; 10 men and 3 women).
All subjects were scanned using an optimized 3D-MPRAGE sequence for enhanced T1 tissue contrast on a 2 Tesla Magnetom scanner (Siemens AG, Munich, Germany).16 Data were analyzed using statistical parametric mapping and optimized VBM (Statistical Parametric Mapping program version 2a [SPM2a], http://www.fil.ion.ucl.ac.uk/spm/software) employing the framework of the general linear model.15-19 Data preprocessing was carried out as follows. First, customized gray and white matter templates were created from the subject structural MRIs. This involved normalizing each structural MRI to the standard statistical parametric map T1 template, segmenting into gray and white matter and cerebrospinal fluid (CSF), and then smoothing each gray and white matter segment with an 8-mm, full width at half maximum, isotropic Gaussian kernel. Subsequently, the images were averaged to create gray and white matter templates in stereotactic space. A fully automated brain extraction procedure was then used to remove nonbrain tissue from the original structural MRIs,17,19 which incorporates a segmentation step. The resulting extracted gray and white matter images were then normalized to the gray and white matter templates, respectively, thus removing any potential contribution of nonbrain voxels. Note that these templates were specific to the present study, created from the participants' own structural scans. The spatial normalization used the residual sum of squared differences as the matching criterion and included both affine transformations and a linear combination of smooth basis functions that modeled global nonlinear shape differences. We then applied the normalization parameters to the original structural images. The rationale for this iterative step was to facilitate optimal segmentation of fully normalized images.
The original segmentation, implicit in the brain extraction step, was performed on affine coregistered images (not in stereotactic space) whereas the Bayesian priors for segmentation were fully normalized (using affine and nonlinear spatial transformations) and were in stereotactic space. Segmentation of fully normalized images (in stereotactic space) is thus optimal. The spatially normalized structural images were resliced to a final voxel size of approximately 1.5 × 1.5 × 1.5 mm3 and optimally segmented into gray and white matter and CSF and non-CSF partitions. Segmentation with SPM2a employs a mixture model cluster analysis (after correcting for nonuniformity in image intensity) to identify voxel intensities matching particular tissue types combined with an a priori knowledge of the spatial distribution of these tissues in normal subjects, derived from probability.
We incorporated an additional step to correct for volume changes introduced during nonlinear spatial transformations. This involved modulating the image partitions (eg, the gray and white matter segments) by the Jacobian determinants derived from the spatial normalization step. In effect, an analysis of modulated data tests for regional differences in the absolute amount (volume) of gray/white matter whereas an analysis of unmodulated data tests for regional differences in concentration of gray/white matter in the voxel.18 Finally, all the normalized, segmented, modulated, and unmodulated images were smoothed with a 12-mm, full width at half maximum, isotropic Gaussian kernel. This conditions the residuals to conform more closely to the Gaussian random field model underlying the statistical process used for adjusting P values.18,19
Global measures of gray matter, white matter, and CSF volumes were calculated from modulated segmented images to test for group differences in overall tissue compartment volumes. Second, separate analytic design matrices were constructed to test for regional differences in gray and white matter tissue compartments between patients with schizophrenia and control subjects. In these analyses, subject data were entered as 4 groups (SZ8, SZ0, CON8, CON0). Age, sex, and total intracranial tissue volume (derived from the sum of the global measures of compartmental volumes) were entered into these analyses as confounding covariates. Thus, we tested for between-group differences that were independent of age, sex, and overall head size.
Statistical parametric maps were constructed to test for morphological differences between subjects with schizophrenia and controls. Data are reported at a significance of P<.05, corrected for multiple comparisons using the false discovery rate correction.14 Note that all these regional effects survived a voxel-wise threshold significance of P< .001, uncorrected for multiple comparisons. We tested first for global differences in tissue compartment volumes. We then tested for regional differences in gray and white matter volumes between all patients with schizophrenia and controls to study the main effect of schizophrenia. Subsequently, we tested for group differences between SZ8 and SZ0 patients and controls in gray matter tissue volume, constraining the analysis to voxels identified as showing a significant main effect of schizophrenia. Where plotted (Figures 2, 3, and 4), the effect size parameter estimates are given in arbitrary units proportional to the percentage difference in volume-adjusted signal. It is the relative differences rather than the scaling of these values that is the key metric.
Figure 2.
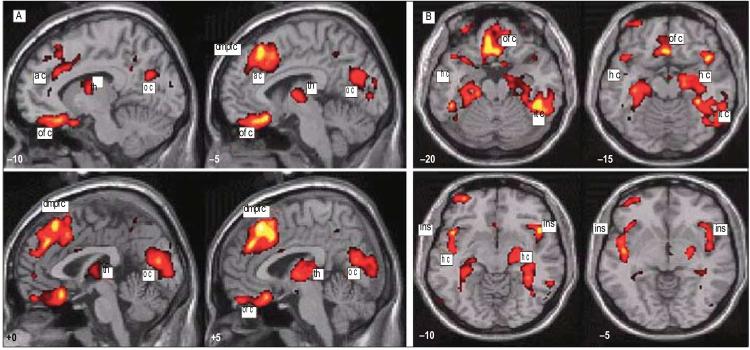
Gray matter differences associated with main effect of schizophrenia. The figure depicts the anatomical locations in red-yellow of significantly (P<.05, corrected) less gray matter volume in groups of patients with schizophrenia (SZ8 and SZ0) relative to matched controls (CON8 and CON0). Data representing group differences are plotted on sagittal (A) and axial (B) sections of a normalized template brain. Distance in millimeters lateral (x coordinate) to the anterior commissure and vertical (z coordinate) to the anterior commissure/posterior commissure line are given at the bottom left corners of these sections. Marked regional foci of gray matter differences are marked as follows: ac indicates anterior cingulate; dmpfc, dorsal medial prefrontal cortex; ofc, orbitofrontal cortex; th,thalamus; oc, occipital cortex; hc, hippocampus; ins, insula; and itc, inferiortemporal lobe.
Figure 3.

Group differences in regional gray matter. Regional gray matter differences between the group of patients with alleles associated with chromosome 8 schizophrenia (SZ8) and their matched controls (CON8) and between the group of patients with schizophrenia but none of the alleles associated with chromosome 8 schizophrenia (SZ0) and their matched controls (CON0). Separate T contrasts were used to test for differences between SZ8 and CON8 and between SZ0 and CON0. These analyses were constrained using masking to regions showing a significant main effect of schizophrenia. Group data are plotted on axial sections of a normalized template brain scan. Regions showing significant differences (P<.05, corrected) between SZ8 and CON8 subjects are depicted in red, and those between SZ0 and CON0 subjects are shown in blue. The axial sections highlight the different patterns of regional gray matter volume deficit associated with these 2 schizophrenic groups; the orbitofrontal deficits in SZ8 patients and the temporal polar and hippocampal deficit in the SZ0 patients are labeled. The distance anterior in millimeters (y coordinate) from the anterior commissure is given in the bottom left corner of each section. ofc indicates orbitofrontal cortex; tp, temporal pole; and hc, hippocampus.
Figure 4.

Between-group gray matter differences in orbitofrontal cortex and temporal pole. A, Orbitofrontal gray matter volume deficit in the group of patients with PCM1 alleles associated with chromosome 8 schizophrenia (SZ8), showing involvement of the olfactory gyrus. Adjacent and to the right are plotted the effect sizes of the different parameter estimates. This is given in arbitrary units proportional to the percentage difference in volume-adjusted signal. It is the relative differences rather than the scaling of these values that is the key metric. A, There was relative reduction in orbitofrontal gray matter (at location x, y, z; −7.5, 34.5, −22.5) in both the SZ8 group and the group of patients with schizophrenia with none of the chromosome 8 PCM1 alleles (SZ0). However, the difference was marked for the SZ8 patients, reaching significance for an interaction, (CON8—SZ8) — (CON0—SZ0). B, Temporal pole gray matter volume deficit in SZ0 patients. Locations of gray matter differences between SZ0 patients and other subjects are illustrated on sagittal and axial sections of a template brain. Adjacent is a plot of parameter estimates for the peak of these differences at the left temporal pole. The greater difference in temporal lobe gray matter volume existed between SZ0 and CON0 subjects. The interaction, (CON0—SZ8)—(CON8—SZ8), was significant. CON8 indicates research subjects who are normal controls matched to the SZ8 cases; CON0, research subjects who are normal controls matched to the SZ0 cases.
RESULTS
GENETIC ASSOCIATION
We reanalyzed the data from our previously published whole genome scan in schizophrenia families using markers on chromosome 8 that were approximately 10 centimorgans apart. The test of association (ETDT) in this family sample between alleles of the marker D8S261 and schizophrenia, but not the other chromosome 8 markers, was found to be significant after taking multiple alleles into account (P= .006, data not shown). We followed this up in the case-control sample and found that D8S26 1 was also significantly associated with schizophrenia (Table 1, empirical P= .009). The linkage disequilibrium relationships between these markers computed with GENECOUNTING/LDPAIRS as in Figure 1 showed that all markers associated with schizophrenia were in significant linkage disequilibrium (P<.001) with 1 or more of the other associated markers (Figure 1). We then genotyped 12 other closely linked markers within the PCM1 gene (Figure 5). Three of these markers also demonstrated significant allelic association. The associated markers were rs445422, rs13276297, and rs370429. The markers D8S2616 and rs3214087 both showed a trend toward association with P values of .07. Markers D8S2615, rs916550, rs412750, rs454755, rs208753, rs3780103, and rs6991775 did not show evidence for association with schizophrenia. The markers showing positive tests of single marker allelic association with schizophrenia are in Table 1. These markers were combined into haplotypes and these showed significant association with schizophrenia as in Table 2. All the associated markers were within or very close to the PCM1 gene and no other genes in the region could be implicated (Figure 5). Empirically derived significance values for these tests of allelic and haplotypic association are in Table 1 and Table 2.
Table 1.
Single Marker Allelic Associations With Schizophrenia in the London-Based Case-Control Sample
| Marker | Distance to Next Marker, bp |
Variant Bases or Marker Fragment Sizes With Allele Frequencies Below | x2 | CLUMP Empirical P Value |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs445422 | 0 | C | T | 5.76 | .02 | ||||||||||||
| Controls | 868 | 12 | |||||||||||||||
| Cases | 758 | 24 | |||||||||||||||
| D8S2616 | 11380 | 213 | 211 | 209 | 207 | 205 | 203 | 201 | 199 | 197 | 195 | 193 | 191 | 189 | 10.09 | .07* | |
| Controls | 1 | 5 | 28 | 57 | 95 | 539 | 42 | 101 | 4 | 5 | 0 | 1 | 0 | ||||
| Cases | 0 | 12 | 26 | 46 | 120 | 457 | 49 | 101 | 5 | 5 | 0 | 0 | 1 | ||||
| rs13276297 | 61 | C | T | 5.50 | .02 | ||||||||||||
| Controls | 840 | 16 | |||||||||||||||
| Cases | 710 | 28 | |||||||||||||||
| D8S261 | 32534 | 128 | 130 | 132 | 134 | 136 | 138 | 140 | 142 | 144 | 146 | 148 | 10.47 | .009† | |||
| Controls | 39 | 266 | 8 | 51 | 308 | 108 | 78 | 10 | 4 | 1 | 13 | ||||||
| Cases | 38 | 206 | 9 | 54 | 297 | 107 | 55 | 28 | 3 | 1 | 12 | ||||||
| rs3214087 | 1037 | Ins | Del | 3.24 | .07 | ||||||||||||
| Controls | 202 | 656 | |||||||||||||||
| Cases | 180 | 720 | |||||||||||||||
| rs370429 | 21 853 | A | G | 6.38 | .01 | ||||||||||||
| Controls | 12 | 856 | |||||||||||||||
| Cases | 25 | 753 | |||||||||||||||
Abbreviation: bp, base pair.
Denotes most likely alleles to be associated with schizophrenia in each haplotype.
Permutation test empirical P value.
Figure 5.
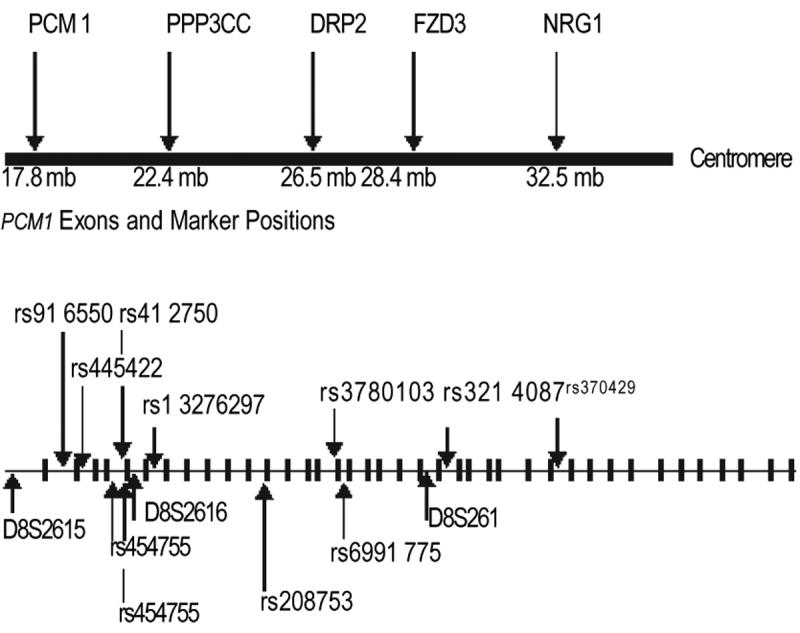
Map of putative chromosome 8p21-22 schizophrenia susceptibility loci on 8p and diagram of the genomic structure of PCM1 showing the relative positions of exons, introns, and genetic markers. Distances are shown in megabases of DNA. PCM1 indicates pericentriolar material 1; PPP3CC, calcineurin; DRP2, dihydropyrimidinase-related protein 2; FZD3, frizzled; and NRG1 , neuregulin.
Table 2.
Haplotypic Associations With Schizophrenia
| Haplotype Label |
No. of Marker Loci |
Alleles Increased in Hap lotypes Showing Association With Schizophrenia |
Frequency in Cases, % |
Frequency in Controls, % |
Global Log Likelihood Ratio x2 |
Permutation Significance |
|---|---|---|---|---|---|---|
| PCM1A1 | 3 Marker | rs445422 allele C, 87366_66 allele T, rs370429 allele G* |
29 | 17 | 14.66 | .003 |
| PCM1 A2 | rs445422 allele T, 87366_66 allele C, rs370429 allele A* |
23 | 12 | |||
| PCM1B1 | 3 Marker | rs454755 allele A, rs3780103alleleG, rs6991 775 allele A* |
40 | 21 | 18.28 | .002 |
| PCM1 B2 | rs454755 allele G, rs37801 03 allele A, rs6991 775 allele A* |
7 | 0 | |||
| PCM1 C1 | 4 Marker | rs454755 allele A, 87366_66 allele C, rs37801 03 allele G, rs6991 775 allele A* |
1 | 0 | 28.72 | .002 |
| PCM1 C2 | rs454755 allele G, 87366_66 allele C, rs37801 03 allele A, rs6991 775 allele A* |
5 | 2 |
Denotes most likely alleles to be associated with schizophrenia in each haplotype.
Two nonoverlapping sets of 3 markers were combined into separate haplotypes (PCM1A1/2 and PCM1B1/2) and all 4 haplotypes showed significant association with schizophrenia (Table 2). Replication of evidence for association with the 3 positive markers was attempted in 2 independent samples. The US replication sample of 100 parent offspring trios including 100 cases of schizophrenia showed that the same dinucleotide allele (142 base pairs [bp]) at the D8S261 locus was also associated with schizophrenia (empirical, P= .03) as in Table 3. A Scottish sample of 200 cases and 200 controls failed to show any significant association between schizophrenia and 3 microsatellite markers (D8S2615, D8S261, D8S2616) near or within the PCM1 locus. Other markers flanking the PCM1 locus, including D8S2615, rs916550, rs412750, rs454755, rs208753, rs3780103, and rs699 1775, showed no individual allelic association with schizophrenia in the London sample. However, markers rs454755, rs3780103, rs6991775, and rs6991775 all showed association with schizophrenia when combined into the haplotypes PCM1A1/2 and PCM1B1/2.
Table 3.
Transmission of Individual Alleles at Marker D8S261 and TDT Analysis in the US Trio Sample*
| Transmission | Dinucleotide Repeat Lengths and Allele Counts | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele size, bp | 130 | 132 | 134 | 136 | 138 | 140 | 142 | 144 | 146 | 148 |
| Allele counts transmitted | 6 | 3 | 27 | 0 | 8 | 33 | 25 | 12 | 6 | 1 |
| Allele counts not transmitted | 4 | 11 | 38 | 3 | 3 | 34 | 10 | 11 | 6 | 1 |
Abbreviations: bp, base pairs; TDT, transmission disequilibrium test.
∼2 9 for allele-wise TDT = 18.88, P = .03. x225 for genotype-wise TDT = 43.21, P = .01.
The research subjects with schizophrenia were selected for the MRI study on the basis that they had inherited alleles associated with schizophrenia at any 2 of the marker loci previously shown to be associated with schizophrenia. These were allele length 142 bp at marker D8S261, allele length 205 bp at marker D8S26 16, allele T at rs445422, allele T at rs13276297, and allele A at rs370429. In analyses of structural MRI scans, patients with schizophrenia had significantly reduced total and relative gray matter volumes compared with controls, and this was apparent in both PCM1-associated cases (SZ8) and non–PCM1-associated (SZ0) patients (mean±SD volumes; controls, 715.8±69 mL; SZ8, 644±63 mL; SZ0, 677±75 mL; F2=5.2, P<.05). Mean±SD percentage gray of gray + white matter volume were controls, 63.9%±0.02%; SZ8, 62.8%±0.01%; and SZ0, 63.3±0.02%; F2=5.2, P<.05. Patients with schizophrenia and controls did not differ in total white matter volume (F2= 1.1, P=.25) or CSF volume (F2=3.0, P=.097). Within the schizophrenic group, we observed no significant difference between SZ8 and SZ0 patient groups in these measures of global tissue volumes.
BRAIN MORPHOLOGY
We examined for group differences in regional brain morphology in separate analyses of gray matter controlling for total intracranial volume, age, and sex. Compared with controls, patients with schizophrenia (combined SZ8 and SZ0 groups) showed a distributed pattern of gray matter volume deficit, encompassing right dorsolateral prefrontal cortex, bilateral dorsomedial and orbitomedial prefrontal cortices, insulae, temporal poles, medial thalamus, posterior hippocampi, inferior temporal (fusiform) cortices, occipital (striate) cortex, and lateral cerebellar hemispheres (Figure 2). Descriptive analysis of trends identified only 1 region in patients with schizophrenia showing a trend in regional gray matter volume augmentation relative to controls. This cluster was located within the primary visual cortex (−3, −59, 3.; P<.001, uncorrected, Z=3.69).
We identified differences between SZ8 and SZ0 patients in the relative distribution of gray matter within those brain areas showing a main effect of diagnosis. Patients in the SZ8 group compared with matched controls had relatively lower gray matter volume, shown in red in Figure 3, located maximally in the bilateral orbitofrontal cortices (P<.05, corrected). In contrast, SZ0 patients had significantly less gray matter, shown in blue in Figure 4, compared with matched controls in temporal poles, particularly on the left, medial temporal lobe (anterior hippocampus), right inferior temporal lobe, medial thalamus, right cerebellum, and dorsomedial prefrontal cortex (P<.05, corrected for main effect of schizophrenia) (Figure 3).
We formally tested for morphometric differences between SZ8 and SZ0 in the pattern of regional gray matter decreases in an interaction analysis in which differences between SZ8 and matched controls were compared with differences between SZ0 and matched controls. The search volume for this analysis, as shown in Figure 4, was constrained using a mask derived from the main effect of diagnosis. Grey matter volume was significantly lower in SZ8 compared with SZ0 patients within medial orbitofrontal cortex bilaterally (x, y, z coordinates [millimeters] from anterior commissure; −7.5, 34.5, −22.5 and 6, 34, −28; P<.05, corrected). Patients in the SZ0 group showed a significant paucity of gray matter compared with SZ8 patients in the left hippocampus (−27, −12, −1), the bilateral temporal poles (−37,21, −42 and 39, 19.5, −34.5), and the right dorsolateral prefrontal cortex (30,46.5, 19.5) (P<.05, corrected) (Figure 1).
COMMENT
The interpretation of the level of statistical significance in the allelic association studies should consider both multiple alleles and multiple markers. In the study of brain morphology, the number of statistical tests carried out should be considered. In the single marker tests of allelic association, we used empirical tests of significance, which do not need further correction for multiple alleles. For the global tests of haplotypic association with schizophrenia, further correction for both multiple alleles and multiple markers are not required because the permutation test significance takes both into account. The finding of extended transmission disequilibrium between D8S261 with schizophrenia in the family linkage sample was the reason to select this and other markers within the PCM1 gene for genotyping in the case-control sample. In addition, the marker D8S26 1 had been independently studied and implicated in schizophrenia in the National Institute of Mental Health trio sample and not as a result of the positive ETDT in the UK family sample or positive association in the UK case-control sample. The other markers showing association in the UK case-control sample were selected for being in linkage disequilibrium with each other. Taking into account that D8S261 had already been implicated in schizophrenia in prior studies and the fact that the additional markers genotyped were not independent tests of association, it can be argued that further corrections of significance values for testing association with multiple markers in the UK case-control sample are not needed.
Our results demonstrate that a chromosomal locus showing replicated evidence of linkage with schizophrenia in several family studies can be identified by observing linkage disequilibrium between markers and schizophrenia in a case-control association sample in which the cases have not been selected for a positive family history of schizophrenia. The fact that we were able to detect linkage disequilibrium in a sample of 450 cases of schizophrenia and find evidence in support of this in an independent sample of 100 US trio cases suggests that the chromosome 8p22 schizophrenia susceptibility alleles may be present in most European populations. As with other reported genetic associations with schizophrenia, it is clear that only a small fraction of research subjects with schizophrenia have a genetic susceptibility from the PCM1 gene locus. This confirms the general findings of linkage studies, which are explicable only by assuming the presence of extensive locus heterogeneity for genes involved in schizophrenia. We attribute the failure to find association in the Scottish sample as being due possiblybecauseonlyalimitedsetof3 markers could be genotyped in this sample. The UK case-control sample employed for this study association to the epsin 4 gene on chromosome 5, which encodes enthoprotin, a clathrin-associated protein involved in vesicle endocytosis in the brain and elsewhere.20
The gene PCM1 is involved in the maintenance of centrosome integrity and the regulation of the microtubule cytoskeleton. Its protein structure bears similarities to the structural myosin proteins, which are microtubule-associated proteins involved in axon guidance, synaptogenesis, functioning of the synapse, and intracellular transport along axons and dendrites.28 It is of note that a different gene disrupted in schizophrenia (DISC1), known to straddle a translocation breakpoint on chromosome 1 that cosegregates with schizophrenia and other types of psychiatric disorder in a single large Scottish pedigree,29 also has similarities to structural proteins such as myosins. It seems likely that PCM1 has a role in the development of the nervous system and neuronal activity. For example, it is known that PCM1 interacts with the brain-specific protein huntingtin-associated protein 1 (HAP1) that binds variably to huntingtin in relation to the number of glutamine repeat lengths present. PCM1 also interacts with cytoskeletal, vesicular, and motor proteins to mediate interactions among these different molecules.28
Our neuroimaging findings further validate the genetic data in so far as we demonstrate distinct differences in regional gray matter abnormalities in patients with chromosome 8p22 PCM1–associated schizophrenia (SZ8) compared with non-chromosome 8 PCMI–associated cases (SZ0). The significance levels reported were corrected for multiple tests and are therefore conservative. A larger sample size, which would reduce the standard error and increase confidence in the results, would require another 450 cases of schizophrenia to be sampled and genotyped at the PCM1 locus to test for association. Therefore, replication in an independent sample is desirable.
Our evidence suggests that brain regions showing the greatest differences between the schizophrenic groups are subsumed within a wider matrix of regional brain abnormalities that span other genetic susceptibilities to schizophrenia. Nevertheless, the peak locations of reduced gray matter volume in SZ8 and SZ0 groups predict important patterns of neuropsychological deficits or vulnerability that can perhaps inform etiopathological mechanisms. Selective gray matter deficit in orbitofrontal cortex, observed in the SZ8 group, is likely to preferentially compromise neural mechanisms supporting reward-related processing and motivational behaviors. In such cases, symptomatology of the patient may be biased toward affective and behavioral features. In contrast, patients with non–chromosome 8 PCMI–associated schizophrenia may exhibit more mnemonic and perceptual features.12 Relevant to this is the recent research showing that families with evidence for linkage to 8p21-22 had significantly more affective deterioration, poorer outcome, more thought disorder, and fewer depressive symptoms than affected individuals from non– 8p21-22–linked families.30
There is evidence of a second schizophrenia susceptibility locus called neuregulin 1 (NRG1) on chromosome 8p21-2231 and 2 replications were successful.32-34 This locus is about 20 million bases toward the centromere from the PCM1 8p22 locus (Figure 5). Gerber et al35 also found evidence of allelic association with schizophrenia on chromosome 8p21 at or near the calcineurin (PPP3CC) gene locus. TwoJapanese research groups now report36,37 association with schizophrenia at the frizzled (FZD3) gene locus suggesting that there may be 4 loci increasing susceptibility on the short arm of chromosome 8. This is compatible with the multiple lod score peaks along the short arm of chromosome 8 in our previous genome linkage scan of schizophrenia.4 In the case-control sample in which we have implicated the PCM1 gene, we have also found weak but significant evidence for association between schizophrenia and the NRG1 gene (Susmitta Datta, PhD, unpublished data, January 2006) but no evidence for association between markers at the PPP3CC or FZD3 genes and schizophrenia (Susmitta Datta, PhD, unpublished data, January 2006). In addition to the epsin 4 gene association, 2 further allelic associations with markers at the FXYD6 (phosphohippolin) gene on chromosome 1 1q23.3 (Khalid Choudhury, BSc, unpublished data, January 2006) and the KIST serine threonine kinase (UHMK1) gene on chromosome 1q23.338 have been found in the same sample. All of these associations have been found by following up localizations originally made in our family linkage analyses.
Further collections of genotyped patients are now needed to confirm the PCM1 8p22 allelic association with schizophrenia and to replicate the finding of brain morphology changes associated with PCM1 -associated marker alleles. Modest but statistically significant levels for PCM1 marker association with schizophrenia have been found in this study. This is compatible with the fact that linkage studies have already proven heterogeneity of linkage in schizophrenia. The linkage studies can only be explicable if one assumes that no single schizophrenia susceptibility gene has an effect in more than 5% to 10% of families. By inference, this would also apply to case-control samples as well. High levels of significance really would not be expected from a case-control sample if only 5% to 10% of cases shared a common genetic susceptibility.
Because some of the single nucleotide polymorphism marker alleles associated with schizophrenia are at both low and high frequencies, sufficient numbers of cases and controls are needed to obtain a good chance of replication. Assuming complete linkage disequilibrium between markers and disease alleles for a significance of P<.05 and a power of 80% of replicating the result (ie, 1 in 4) with a low-frequency allele present in 5% of controls and 10% of cases, then 600 cases and 600 controls should be enough. For a high-frequency disease-associated allele present in 50% of cases and in 45% of controls, then 1300 cases and 1300 controls are needed. We can now sequence DNA from cases who have inherited PCM1 gene marker alleles associated with schizophrenia to define the precise etiological base pair changes or mutations increasing susceptibility to schizophrenia. If these can be found, then exactly how PCM1 shapes regional brain structure and function can be understood. The alternative splicing of PCM1 messenger RNA, interaction of PCM1 with other proteins, posttranslational modification, phosphorylation, and glycosylation may all play a role in mediating genetic effects on the disease.
References
- 1.Gurling HMD. The genetics of the schizophrenias. Genetics of Mental Disorders, Part II: Clinical Issues. In: Papadimitriou GN, Mendlewicz J, editors. Balliere's Clinical Psychiatry, International Practice and Research. Vol. 2. 1996. pp. 15–46. [Google Scholar]
- 2.Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, Ullrich G, McGrath J, Kasch L, Lamacz M, Thomas MG, Gehrig C, Radhakrishna U, Snyder SE, Balk KG, Neufeld K, Swartz KL, DeMarchi N, Papadimitriou GN, Dikeos DG, Stefanis CN, Chakravarti A, Childs B, Housman DE, Kazazian HH, Antonarakis S, Pulver AE. Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet. 1998;20:70–73. doi: 10.1038/1734. [DOI] [PubMed] [Google Scholar]
- 3.Brzustowicz LM, Honer WG, Chow EWC, Little D, Hogan J, Hodgkinson K, Bassett AS. Linkage of familial schizophreniato chromosome 13q32. Am J Hum Genet. 1999;65:1096–1103. doi: 10.1086/302579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet. 2001;68:661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kendler KS, MacLean CJ, O'Neill FA, Burke J, Murphy B, Duke F, Shinkwin R, Easter SM, Webb BT, Zhang J, Walsh D, Straub RE. Evidence for a schizophreniavulnerability locus on chromosome 8p in the Irish Study of High-Density Schizophrenia Families. Am J Psychiatry. 1996;153:1534–1540. doi: 10.1176/ajp.153.12.1534. [DOI] [PubMed] [Google Scholar]
- 6.Blaveri E, Kalsi G, Lawrence J, Quested D, Moorey H, Lamb G, Kohen D, Shiwach R, Chowdhury U, Curtis D, McQuillin A, Gramoustianou ES, Gurling HM. Genetic association studies of schizophrenia using the 8p21-22 genes: prepronociceptin (PNOC), neuronal nicotinic cholinergic receptor alpha polypeptide 2 (CHRNA2) and arylamine N-acetyltransferase 1 (NAT1) Eur J Hum Genet. 2001;9:469–472. doi: 10.1038/sj.ejhg.5200646. [DOI] [PubMed] [Google Scholar]
- 7.Weinberger DR, Torrey EF, Neophytides AN, Wyatt RJ. Structural abnormalities in the cerebral cortex of chronic schizophrenic patients. Arch Gen Psychiatry. 1979;36:935–939. doi: 10.1001/archpsyc.1979.01780090021002. [DOI] [PubMed] [Google Scholar]
- 8.Weinberger DR. Computed tomography (CT) findings in schizophrenia: speculation on the meaning of it all. J Psychiatr Res. 1984;18:477–490. doi: 10.1016/0022-3956(84)90035-9. [DOI] [PubMed] [Google Scholar]
- 9.Harvey I, Ron MA, Du Boulay G, Wicks D, Lewis SW, Murray RM. Reduction of cortical volume in schizophrenia on magnetic resonance imaging. Psychol Med. 1993;23:591–604. doi: 10.1017/s003329170002537x. [DOI] [PubMed] [Google Scholar]
- 10.Lim KO, Tew W, Kushner M, Chow K, Matsumoto B, DeLisi LE. Cortical gray matter volume deficit in patients with first-episode schizophrenia. Am J Psychiatry. 1996;153:1548–1553. doi: 10.1176/ajp.153.12.1548. [DOI] [PubMed] [Google Scholar]
- 11.Wright IC, Rabe-Hesketh S, Woodruff PW, David AS, Murray RM, Bullmore ET. Meta-analysis of regional brain volumes in schizophrenia. Am J Psychiatry. 2000;157:16–25. doi: 10.1176/ajp.157.1.16. [DOI] [PubMed] [Google Scholar]
- 12.Liddle PF. Schizophrenic syndromes, cognitive performance and neurological dysfunction. Psychol Med. 1987;17:49–57. doi: 10.1017/s0033291700012976. [DOI] [PubMed] [Google Scholar]
- 13.Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston K, Evans AC. A unified statistical approach for determining significant signals in images of cerebral activation. Human Brain Mapping. 1996;4:58–73. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 14.Genovese CR, Lazar NA, Nichols T, Tew W. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 15.Deichmann R, Good CD, Turner RF. Inhomogeneity compensation in structural brain imaging. Magn Reson Med. 2002;47:398–402. doi: 10.1002/mrm.10050. [DOI] [PubMed] [Google Scholar]
- 16.Deichmann R, Good CD, Josephs O, Ashburner J, Turner R. Optimization of 3-DMP-RAGE sequences for structural brain imaging. Neuroimage. 2000;12:112–127. doi: 10.1006/nimg.2000.0601. [DOI] [PubMed] [Google Scholar]
- 17.Ashburner J, Friston KJ. Why voxel-based morphometry should be used. Neuroimage. 2001;14:1238–1243. doi: 10.1006/nimg.2001.0961. [DOI] [PubMed] [Google Scholar]
- 18.Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 19.Ashburner J, Friston KJ. Voxel-based morphometry: the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 20.Pimm J, McQuillin A, Thirumalai S, Lawrence J, Quested D, Bass N, Lamb G, Moorey H, Datta SR, Kalsi G, Badacsonyi A, Kelly K, Morgan J, Punukollu B, Curtis D, Gurling H. The epsin 4 gene on chromosome 5q, which encodes the clathrin associated protein enthoprotin, is involved in the genetic susceptibility to schizophrenia. Am J Hum Genet. 2005;76:902–907. doi: 10.1086/430095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endicott J, Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry. 1978;35:837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 23.Sham PC, Curtis D. An extended transmission/disequilibrium test (TDT) for multi-allele marker loci. Ann Hum Genet. 1995;59:323–336. doi: 10.1111/j.1469-1809.1995.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 24.Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet. 1995;59:97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhao JH, Curtis D, Sham PC. Model-free analysis and permutation tests for allelic associations. Hum Hered. 2000;50:133–139. doi: 10.1159/000022901. [DOI] [PubMed] [Google Scholar]
- 26.Zhao JH, Lissarrague S, Essioux L, Sham PC. GENECOUNTING: haplotype analysis with missing genotypes. Bioinformatics. 2002;18:1694–1695. doi: 10.1093/bioinformatics/18.12.1694. [DOI] [PubMed] [Google Scholar]
- 27.Terwilliger JD, Ott J. Handbook for Human Genetic Linkage. Baltimore, Md: Johns Hopkins University Press; 1994. [Google Scholar]
- 28.Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, Holzbaur EL, Ross CA. Huntingtin-associated protein 1 (HAP1) interacts with the p150 Glued subunit of dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 29.Millar JK, Christie S, Anderson S, Lawson D, Hsiao-Wei Loh D, Devon RS, Arveiler B, Muir WJ, Blackwood DH, Porteous DJ. Genomic structure and localisation within a linkage hotspot of Disrupted In Schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol Psychiatry. 2001;6:173–178. doi: 10.1038/sj.mp.4000784. [DOI] [PubMed] [Google Scholar]
- 30.Kendler KS, Myers JM, O'Neill FA, Martin R, Murphy B, MacLean CJ, Walsh D, Straub RE. Clinical features of schizophrenia and linkage to chromosomes 5q, 6p, 8p, and 10p in the Irish Study of High-Density Schizophrenia Families. Am J Psychiatry. 2000;157:402–408. doi: 10.1176/appi.ajp.157.3.402. [DOI] [PubMed] [Google Scholar]
- 31.Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, Gunnarsdottir S, Walker N, Petursson H, Crombie C, Ingason A, Gulcher JR, Stefansson K, Clair D. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72:83–87. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang JZ, Si TM, Ruan Y, Ling YS, Han YH, Wang XL, Zhou M, Zhang HY, Kong QM, Liu C, Zhang DR, Yu YQ, Liu SZ, Ju GZ, Shu L, Ma DL, Zhang D. Association study of neuregulin 1 gene with schizophrenia. Mol Psychiatry. 2003;8:706–709. doi: 10.1038/sj.mp.4001377. [DOI] [PubMed] [Google Scholar]
- 34.Iwata N, Suzuki T, Ikeda M, Kitajima T, Yamanouchi Y, Inada T, Ozaki N. No association with the neuregulin 1 haplotype to Japanese schizophrenia. Mol Psychiatry. 2004;9:126–127. doi: 10.1038/sj.mp.4001456. [DOI] [PubMed] [Google Scholar]
- 35.Gerber DJ, Hall D, Miyakawa T, Demars S, Gogos JA, Karayiorgou M, Tonegawa S. Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin gamma subunit. Proc Natl Acad Sci U S A. 2003;100:8993–8998. doi: 10.1073/pnas.1432927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katsu T, Ujike H, Nakano T, Tanaka Y, Nomura A, Nakata K, Takaki M, Sakai A, Uchida N, Imamura T, Kuroda S. The human frizzled-3 (FZD3) gene on chromosome 8p21, a receptor gene for Wnt ligands, is associated with the susceptibility to schizophrenia. Neurosci Lett. 2003;353:53–56. doi: 10.1016/j.neulet.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Si T, Ling Y, Ruan Y, Han Y, Wang X, Zhang H, Kong Q, Li X, Liu C, Zhang D, Zhou M, Yu Y, Liu S, Shu L, Ma D, Wei J, Zhang D. Association study of the human FZD3 locus with schizophrenia. Biol Psychiatry. 2003;54:1298–1301. doi: 10.1016/s0006-3223(03)00291-9. [DOI] [PubMed] [Google Scholar]
- 38.Puri V, McQuillin A, Choudhury K, Datta S, Pimm J, Thirumalai S, Krasucki R, Lawrence J, Quested D, Bass N, Moorey H, Morgan J, Punukollu B, Kandasami G, Curtis D, Gurling H. Fine mapping by genetic association implicates the chromosome 1q23.3 gene UHMK1, encoding a serine/threonine protein kinase, as a novel schizophrenia susceptibility gene. Biol Psychiatry. 2006 doi: 10.1016/j.biopsych.2006.06.014. In press. [DOI] [PubMed] [Google Scholar]