Abstract
Auxin is an essential regulator of plant organogenesis. Most key genes in auxin biosynthesis, transport, and signaling belong to gene families, making it difficult to conduct genetic analysis of auxin action in plant development. Herein we report the functional analysis of several members of 2 gene families (NPY/ENP/MAB4 genes and AGC kinases) in auxin-mediated organogenesis and their relationships with the YUC family of flavin monooxygenases that are essential for auxin biosynthesis. We show that 5 NPY genes (NPY1 to NPY5) and 4 AGC kinases (PID, PID2, WAG1, and WAG2) have distinct, yet overlapping, expression patterns. Disruption of NPY1 does not cause obvious defects in organogenesis, but npy1 npy3 npy5 triple mutants failed to make flower primordia, a phenotype that is also observed when AGC kinase PID is compromised. Inactivation of YUC1 and YUC4 in npy1 background also phenocopies npy1 npy3 npy5 and pid. Simultaneous disruption of PID and its 3 closest homologs (PID2, WAG1, and WAG2) completely abolishes the formation of cotyledons, which phenocopies npy1 pid double mutants and yuc1 yuc4 pid triple mutants. Our results demonstrate that NPY genes and AGC kinases define 2 key steps in a pathway that controls YUC-mediated organogenesis in Arabidopsis.
Keywords: NPY1/ENP1/MAB4, YUC, PINOID, embryogenesis, cotyledon
Formation of embryonic and postembryonic organs is an essential process for normal plant development and is regulated by intrinsic signals and environmental cues. Genetic analyses of Arabidopsis mutants with defects in organogenesis demonstrated that the plant hormone auxin plays a key role in determining the formation and patterning of lateral organs. Disruption of either auxin biosynthesis (1, 2) or polar auxin transport/auxin signaling (3–5) leads to defects in embryogenesis and in the formation of leaves and flowers. Auxin has been proposed as a morphogen that provides instructive signals for the formation of organs (6–9). The current model of organogenesis in Arabidopsis is that an auxin maximum (auxin peak) at the flanks of the apical meristem is necessary and sufficient to initiate the formation of lateral organs (9, 10). However, the exact mechanisms by which auxin regulates organogenesis are not fully resolved.
It is still not understood how auxin maxima are generated and maintained. Much of the work in this aspect in the past decade was centered on the active transport of auxin mediated by the PIN-FORMED (PIN) auxin efflux carriers and AUXIN1 (AUX1) influx carriers (4, 5, 10). Computer-assisted modeling on auxin dynamics and organogenesis based on PIN protein localization further improved our understanding of how auxin transport may contribute to the formation of auxin peaks (9, 11). However, recent progress in auxin biosynthesis reveals a more complicated picture. It appears that both local auxin production and polar auxin transport contribute to the creation and maintenance of auxin peaks. Mutations in the auxin efflux carrier PIN1 disrupt the initiation of floral organs (5). Simultaneously inactivation of YUCCA1 (YUC1) and YUC4, which encode homologous flavin-containing monooxygeases essential for de novo auxin biosynthesis (1, 12), also leads to defects in flower development (1). The yuc1 yuc4 pin1 triple mutants fail to make any true leaves (2), a phenotype not observed in either pin1 or yuc1 yuc4 alone, demonstrating that leaf initiation is controlled by both the auxin biosynthetic YUC genes and the auxin transport PIN genes. Furthermore, the auxin influx carrier mutant aux1 itself does not show any defects in organogenesis in the aerial parts of Arabidopsis, but aux1 fails to make flowers in the yuc1 yuc2 yuc4 yuc6 quadruple-mutant background, indicating that the functions of AUX1 in organogenesis are masked by local auxin biosynthesis (2).
One of the difficulties in conducting genetic analysis of auxin pathways is that almost all of the key components in auxin biosynthesis, polar transport, and auxin signaling belong to gene families whose members have overlapping functions. For example, the 11 YUC flavin monooxygenases in Arabidopsis catalyze a rate-limiting step in auxin biosynthesis and disruption of a single YUC gene does not cause any obvious developmental defects, but some double- and triple-mutant combinations have severe defects in development (1, 12). For the PIN family of efflux carriers and the auxin response factor (ARF) family, inactivation of PIN1 or MONOPTEROS (MP)/ARF5 alone is sufficient to cause dramatic developmental defects (5, 13, 14). However, it has been demonstrated that PIN1 has overlapping functions with other PIN genes (4) and that MP has overlapping functions with ARF7 and ARF19 (15). Such genetic complexities in auxin pathways make it difficult to define the functions of an individual of a gene family and conduct epistasis analysis of the auxin mutants because inactivation of one gene does not lead to a complete null of the gene function because of the compensatory effects from the other homologous genes.
To further elucidate the molecular mechanisms by which auxin regulates plant organogenesis, we conducted a genetic screen for enhancers of yuc1 yuc4 double mutants on the basis of the hypothesis that the yuc1 yuc4 double mutants provide a sensitized background for identifying novel components that are involved in auxin-regulated organogenesis. Our previous studies have demonstrated that the yuc mutants synergistically interact with polar auxin transport mutants (2). We focused our attention to mutants that fail to make flowers, but still develop an inflorescence in the yuc1 yuc4 background. Such naked inflorescences without flowers are called pin-like inflorescences. Formation of pin-like inflorescences has become a hallmark for malfunction of auxin pathways because known pin-like mutants such as pin1 (5), pinoid (pid) (16, 17), and mp (14) all are involved in aspects of auxin biology.
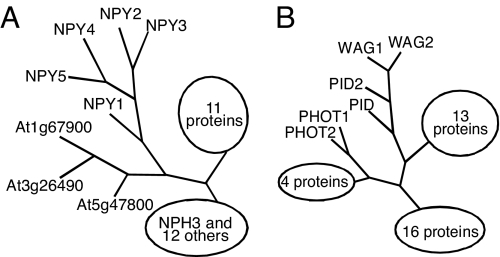
We identified a yuc1 yuc4 enhancer naked pins in yuc mutants 1 (npy1), which forms pin-like inflorescences in the yuc1 yuc4 background, but not in wild-type background (18). Mutant npy1 is allelic to enhancer of pinoid 1 (enp1) and macchi-bou 4 (mab4) (19, 20), which failed to develop cotyledons in the pid background. NPY1/ENP1/MAB4 encodes a plant-specific protein that contains a BTB (Bric-a-brac, Tramtrack, Broad-complex) domain at the N-terminal region and an NPH3 (NON-PHOTOTROPIC HYPOCOTYL 3) domain in the middle. NPY1 belongs to a large gene family with 32 members in the Arabidopsis genome (Fig. 1A). The founding member of this family, NPH3, mediates phototropic response downstream of the photoreceptor PHOT1 (21). Both PHOT1 and PID are Ser/Thr kinases that belong to the AGC kinase superfamily (Fig. 1B). The AGC kinases are the collective name for cAMP-dependent protein kinase A, cGMP-dependent protein kinase G, and phospholipids-dependent protein kinase C (22). The fact that NPY1 is homologous to NPH3 and PID is homologous to PHOT1 suggests that auxin-regulated organogenesis and phototropic responses are analogous. Furthermore, both processes require the involvement of an auxin response factor. Inactivation of NPH4/ARF7 leads to defects in phototropic response and disruption of MP/ARF5 leads to the formation of pin-like inflorescences (14, 23). Analysis of genetic interactions among yuc1 yuc4, npy1, and pid has put the genes in a genetic context in regulating Arabidopsis organogenesis (18).
Fig. 1.
Schematic trees of NPY proteins and AGC kinases. (A) A tree of NPY proteins that belong to a family with 32 members in Arabidopsis. The founding member of this superfamily is NPH3. For simplicity, the details of the NPH3 clade and the other large clade are not shown. (B) A tree of the AGC kinase family. More detailed phylogenetic trees of the 2 families have been published (18, 21, 24). The GenBank accession numbers for NPY genes are At4g31820 (NPY1), At2g14820 (NPY2), At5g67440 (NPY3), At2g23050 (NPY4), and At4g37590 (NPY5). The GenBank accession numbers for PID and its homologs are At2g34650 (PID), At2g26700 (PID2), At1g53700 (WAG1), and At3g14370 (WAG2).
The npy1 pid double mutants and the yuc1 yuc4 pid triple mutants failed to make cotyledons, a phenotype that was not observed in pid, npy1, or yuc1 yuc4 alone (18). The synergistic genetic interaction can be explained as the genes are involved in parallel pathways. Alternatively, it can also be interpreted that the YUCs, NPY1, and PID are in a linear pathway, providing that each gene has redundant partners in the Arabidopsis genome. To distinguish between the 2 possibilities, it is necessary to analyze whether other NPY1-like genes and PID-like genes also participate in auxin-regulated plant development. Herein we demonstrate that 2 closest NPY1 homologs in Arabidopsis also play an important role in the formation of flowers. Inactivation of one or more NPY1 homologs in npy1 background leads to the formation of pin-like inflorescences, a phenotype observed in pid and yuc1 yuc4 npy1 triple mutants (18). Furthermore, we show that inactivation of the 3 closest PID homologs in the pid background completely abolishes the formation of cotyledons, a phenotype that is also observed in yuc1 yuc4 pid triple mutants (18) and npy1 pid double mutants (18, 19). Our analyses establish that members of YUC, NPY1, and PID families participate in a linear pathway in regulating auxin-mediated organogenesis in Arabidopsis and NPY genes and PID-like AGC kinases define 2 essential steps in the pathway. This study also provides a model for conducting genetic analysis on a network of multiple gene families.
Results
NPY Genes Have Unique and Overlapping Expression Patterns.
Mutations in NPY1 greatly enhanced the phenotypes of yuc1 yuc4 double mutants (18) and pid mutants (18, 19), but npy1 itself does not have dramatic developmental defects. It is likely that a subset of the NPY1 homologs has at least partially overlapping functions with NPY1. On the basis of sequence homology, the 32 members of the NPY1/NPH3 family in Arabidopsis can be divided into 4 subgroups: the NPH3/RPT2 group, the NPY group, and 2 other groups. In this article we focus our effort on the NPY group that includes 5 members named as NPY1 to NPY5 (Fig. 1A). Detailed phylogenetic analyses of the NPY1/NPH3 family and the AGC kinases have been described (18, 21, 24).
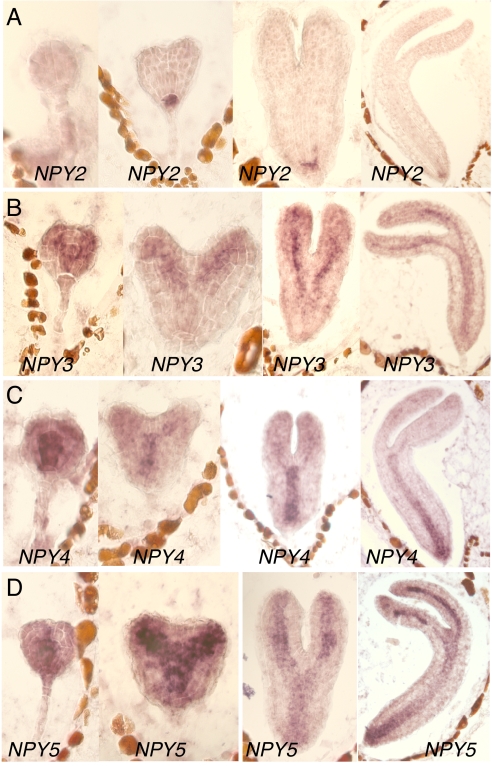
First, we analyzed the expression patterns of the 5 NPY genes by RNA in situ hybridization. It was evident that all 5 NPY genes were expressed during embryogenesis (Fig. 2) (18), but with distinct, yet overlapping, patterns. NPY1 was expressed mainly in the apical regions of embryos including cotyledon tips and the apical meristem (18). In contrast, NPY2 was specifically expressed in the hypophysis and the root meristems in the embryos (Fig. 2A). The expression of NPY3 was concentrated in provascular and vascular systems (Fig. 2B). Both NPY4 and NPY5 were also expressed in the cells that would differentiate into vascular bundles, but NPY4 and NPY5 appeared to have different tissue specificities (Fig. 2 C and D). NPY4 was expressed mainly in the hypocotyls (Fig. 2C), whereas NPY5 was more concentrated in cotyledons (Fig. 2D). Overall, the expression of the 5 NPY genes marked the 2 meristems, provascular tissues, and the cotyledon tips during embryogenesis.
Fig. 2.
The expression patterns of NPY genes during embryogenesis. (A) NPY2. (B) NPY3. (C) NPY4. (D) NPY5. The expression of the NPY genes were detected by using RNA in situ hybridization.
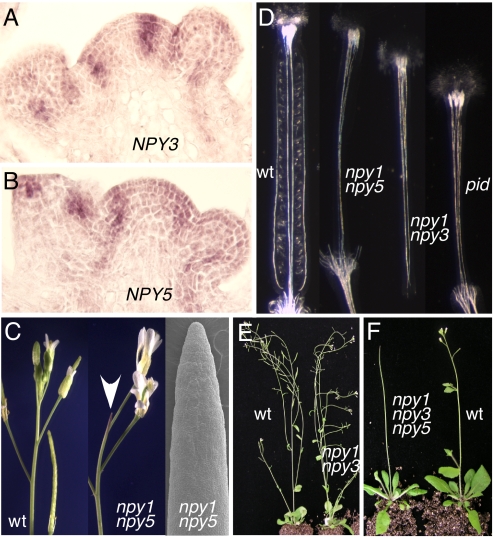
In the inflorescence apex, NPY1 was expressed in the apical meristem and flower primordia (18). In apical meristem, the NPY1 expression appeared to be restricted to the L1 layer (18). We did not detect any expression of NPY2 in the inflorescence apex by RNA in situ hybridization (data not shown), which is consistent with the idea that NPY2 may play a more specific role in root development. NPY3 expression was not detected in the apical meristem; instead it marked the incipient sites of new floral primordia (Fig. 3A). NPY3 mRNA was also detected in the early stages of flowers (Fig. 3A). NPY4 was weakly expressed in the vascular tissues of inflorescences apex (data not shown). Among the 4 NPY1 homologs, the expression pattern of NPY5 was the most similar to that of NPY1 (Fig. 3B) (18). NPY5 mRNA was detected in the apical meristem and young flowers (Fig. 3B). However, NPY5 showed a more pronounced expression in the incipient sites of flower primordia than NPY1. Overall, the expression patterns of NPY5 overlapped greatly with those of NPY1 and NPY3 in the inflorescence apex.
Fig. 3.
The roles of NPY genes in Arabidopsis development. (A) Expression of NPY3 in the inflorescence apex. (B) NPY5 expression in the inflorescence apex. (C) Disruption of NPY1 and NPY5 led to the formation of pin-like inflorescences. A scanning electron microscope micrograph of npy1 npy5 inflorescence is shown at right. The arrow points to a pin-like inflorescence. (D) NPY genes were required for proper gynoecium development. WT gynoecium had well connected and defined median and lateral vascular bundles. The ovules and valve tissues were also evident. However, npy1 npy5, or npy1 npy3, or pid all failed to make proper valves and developed abnormal vascular tissues in gynoecia. From left to right, WT, npy1 npy5, npy1 npy3, and pid. (E) The npy1 npy3 double mutants were sterile. (F) The triple mutants of npy1 npy3 npy5 formed strong pin-like inflorescences.
Inactivation of NPY Genes Leads to the Formation of Pin-Like Inflorescences.
We isolated T-DNA insertion mutants of NPY1 and its 4 closest homologs (Fig. S1). None of the single mutants displayed any obvious defects in organogenesis except npy1, which had subtle defects in cotyledon development (18). The single npy mutants never developed pin-like inflorescences (data not shown). However, when npy1 and npy5 were combined, the resulting double mutants had pin-like inflorescences (Fig. 3C), a phenotype that was also observed in pin1 (5), pid, mp, and npy1 yuc1 yuc4 triple mutants. The npy1 npy5 double mutants still made some flowers, but the flowers were abnormal (Fig. 3C). Like the flowers in pid and pin1, the npy1 npy5 flowers often contained multiple petals (Fig. S2). The gynoecium of npy1 npy5 often lacked valves and displayed vascular defects (Fig. 3D), which were also observed in pid and pin1 mutants. The observed phenotypes of npy1 npy5 correlated well with the fact that both NPY1 and NPY5 were expressed in the inflorescence meristem and that the expression patterns of the 2 genes overlapped (Fig. 3) (18).
The double mutants npy1 npy3 also displayed defects in flower development (Fig. 3E). Inactivation of both NPY1 and NPY3 led to sterile plants (Fig. 3E). The flowers in npy1 npy3 failed to develop proper valves and vascular tissues in gynoecium, phenotypes not observed in either npy1 or npy3 alone (Fig. 3D). The defects in gynoecium development in npy1 npy3 double mutants were very similar to those observed in npy1 npy5 and pid (Fig. 3D). The genetic enhancement of npy1 by npy3 indicated that NPY1 and NPY3 also had overlapping functions, which is consistent with the expression patterns of NPY1 and NPY3 (Fig. 3).
Interestingly, the double mutants of npy3 npy5 did not display any obvious developmental defects, suggesting that NPY1 plays a more prominent role. We generated all other possible double mutant combinations including npy1 npy2 and npy1 npy4, but only npy1 npy3 and npy1 npy5 displayed obvious developmental defects. The triple mutants of npy1 npy3 npy5 had stronger phenotypes than the double mutants npy1 npy3 or npy1 npy5 (Fig. 3F). The triple mutants made very few flowers and developed strong pin-like inflorescences (Fig. 3F). We further inactivated NPY2 and NPY4 in the npy1 npy3 npy5 triple-mutant background; the resulting quintuple mutants were very similar to the npy1 npy3 npy5 triple mutants.
Three PID Homologs Were Expressed During Embryogenesis.
PID is expressed during Arabidopsis embryogenesis, and PID has been shown to play important roles in cotyledon development. Because both npy1 pid and yuc1 yuc4 pid failed to develop cotyledons while pid alone made cotyledons, we hypothesized that the PID homologs and PID may have overlapping functions in cotyledon development. In this article, we focus on the 3 closest PID homologs (PID2, WAG1, and WAG2) (Fig. 1B) and their roles in plant development. Both WAG1 and WAG2 were shown to play a role in root development and gravitropic response (25), whereas the role of PID2 [previously called AGC1-10 or AGC3-4 (22, 24)] in plant development has not been analyzed.
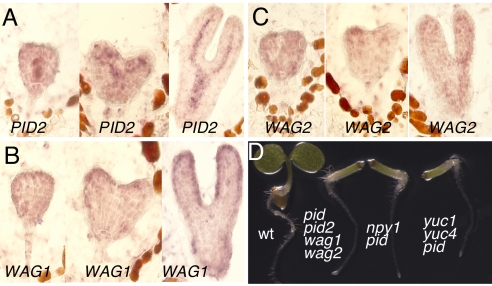
We first used RNA in situ hybridization to investigate whether the PID homologs are expressed during embryogenesis. We detected the mRNAs of the 3 PID homologs during embryogenesis (Fig. 4). Both WAG1 and WAG2 were expressed throughout the embryogenesis; higher expression of WAG1 and WAG2 in the cotyledon primordia at heart stages was evident (Fig. 4). Compared with WAG1 and WAG2, PID2 had much higher expression (Fig. 4). The PID2 mRNA was restricted mainly in provascular tissues (Fig. 4), a pattern that was very similar to those of NPY3, NPY4, and NPY5 (Fig. 2).
Fig. 4.
The expression patterns of the 3 closest homologs of PID and their roles in embryogenesis. (A) PID2 expression. (B) WAG1 expression. (C) Expression pattern of WAG2; the expression patterns were revealed by in situ RNA hybridization. (D) Essential roles of PID and its homologs in cotyledon development. The quadruple mutants pid pid2 wag1 wag2 failed to make cotyledons. The same phenotypes were also observed in npy1 pid and yuc1 yuc4 pid. From left to right, WT, pid pid2 wag1 wag2 quadruple mutants, npy1 pid double mutants, and yuc1 yuc4 pid triple mutants.
PID and Its Homologs Are Essential for the Formation of Cotyledons.
We isolated T-DNA insertion mutants of PID, PID2, WAG1, and WAG2 (Fig. S3). Inactivation of PID led to the formation of 3 cotyledons, but the phenotype was not fully penetrant. Single mutants of pid2, wag1, and wag2 did not display any obvious defects in embryogenesis (data not shown). When we put pid2 in pid background, the resulting double mutants behaved like pid during embryogenesis (Table 1). Most seedlings (72%) of the double mutants pid wag1 had normal-looking cotyledons, but we observed some seedlings (24%) with only residual cotyledons or no cotyledon at all (1%) (Table 1) (Fig. S4). Like pid wag1, ≈5% of pid wag2 had residual cotyledons, 14% had no cotyledons, and 81% appeared normal. Other double mutants without pid such as wag1 wag2 did not show obvious embryonic defects.
Table 1.
Genetic analyses of mutant combinations of pid, pid2, wag1, and wag2
| Parent genotype | Mutant genotype | Mutant genotype analysis |
Mutant phenotype analysis |
||||
|---|---|---|---|---|---|---|---|
| No. of seedlings (% of total seedlings genotyped) |
No. of seedlings (% of total number of mutant seedlings) |
||||||
| Expected | Observed | Seedlings genotyped | With cotyledons | Residual cotyledons | Without cotyledons | ||
| pid+/−pid2 | pid pid2 | 36 (25) | 33 (23) | 144 | 33 (100) | 0 (0) | 0 (0) |
| Pid+/−wag1 | pid wag1 | 27 (25) | 25 (23) | 109 | 18 (72) | 6 (24) | 1 (4) |
| pid+/−wag2 | pid wag2 | 40 (25) | 37 (23) | 160 | 30 (81) | 2 (5) | 5 (14) |
| pid+/−pid2 wag1+/− | pid pid2 wag1 | 10 (6) | 13 (8) | 164 | 10 (77) | 1 (8) | 2 (15) |
| pid+/−pid2+/−wag2 | pid pid2 wag2 | >9 (>6)* | 26 (18) | 145 | 19 (73) | 3 (12) | 4 (15) |
| pid+/−wag1+/−wag2 | pid wag1 wag2 | 8 (6) | 7 (6) | 121 | 0 (0) | 0 (0) | 7 (100) |
| pid+/−wag1 wag2+/− | pid wag1 wag2 | 8 (6) | 5 (4) | 135 | 0 (0) | 0 (0) | 5 (100) |
| pid+/−wag1 wag2 | pid wag1 wag2 | 29 (25) | 30 (26) | 116 | 2 (7) | 3 (10) | 25 (83) |
| pid+/−wag1 wag2+/− | pid wag1 wag2+/− | 17 (13) | 12 (9) | 135 | 1(8) | 4 (33) | 7 (58) |
| pid+/−wag1+/−wag2 | pidwag1+/−wag2 | 15 (13) | 18 (15) | 121 | 2 (11) | 3 (17) | 13 (72) |
| pid2 wag1 wag2 | pid2 wag1 wag2 | 24 (100) | 24 (100) | 24 | 24 (100) | 0 (0) | 0 (0) |
| pid+/−pid2 wag1 wag2 | pid pid2 wag1wag2 | 59 (25) | 59 (25) | 237 | 1 (2) | 0 (0) | 58 (98) |
*Both PID and PID2 are located on chromosome II and are linked.
We also analyzed the triple-mutant combinations of pid, pid2, wag1 and wag2 (Table 1). Whereas pid2 wag1 wag2 did not show any obvious defects, the other triple mutants displayed defects in cotyledon development. The majority of pid pid2 wag1 and pid pid2 wag2 had normal cotyledons (77% and 73%, respectively), and a small number of the triple mutants had cotyledon defects (Table 1). Unlike the other triple mutants, most of the pid wag1 wag2 triple mutants (88%) failed to make any cotyledons and only 7% had residue cotyledons (Table 1). Less than 5% of pid wag1 wag2 displayed normal cotyledons. We noticed that heterozygous wag1 or wag2 increased the frequency of cotyledon defects in pid wag2 and pid wag1, respectively (Table 1). The no-cotyledon phenotype in pid wag1 wag2 was identical to that of npy1 pid and yuc1 yuc4 pid (Fig. 4). The failure to make cotyledons in pid wag1 wag2 took place during embryogenesis (Fig. S4).
We further analyzed the pid pid2 wag1 wag2 quadruple mutants. Among the 59 seedlings of the quadruple mutants, 58 seedlings had no cotyledons, suggesting the phenotype was almost fully penetrant (Table 1). Our genetic analyses indicate that PID plays a more prominent role than the PID homologs during embryogenesis. Whereas the majority of pid wag1 wag2 did not make cotyledons (88%), the frequency was greatly increased (98%) in pid pid2 wag1 wag2.
Discussion
Herein we present direct evidence that the NPY genes play an essential role in Arabidopsis organogenesis. Our genetic analysis of NPY genes and PID genes demonstrates that each gene family represents a key step in exerting the functions of auxin during Arabidopsis development.
NPY Genes Collectively Control Organogenesis in Arabidopsis.
NPY1 belongs to a large gene family (Fig. 1A). Genetic screens for enhancers of yuc1 yuc4 and pid led to the discovery of NPY1 as a key component in auxin-regulated organogenesis. The npy1 pid double mutants failed to develop cotyledons and the npy1 yuc1 yuc4 formed pin-like inflorescences (18), but npy1 alone did not display obvious developmental defects. In addition to the interpretation that NPY1 works in the same pathway as PID and YUC genes, the observed genetic enhancements among yuc, pid, and npy1 can be accounted for if NPY1 and PID function in parallel pathways. Inactivation of either NPY3 or NPY5 in the npy1 background led to dramatic flower defects similar to those observed in pid (Fig. 3). Furthermore, the npy1 npy3 npy5 display strong pin-like phenotypes (Fig. 3), which is identical to those of strong pid alleles, demonstrating that NPY1 and PID are unlikely to function in 2 parallel pathways.
NPY1, NPY3, and NPY5 displayed overlapping and distinct expression patterns (Figs. 2 and 3), suggesting that each gene may also have unique functions. It is difficult to dissect the unique functions of the genes in a family. It may require us to put individual mutants in a sensitized background. For example, YUC1 and YUC4 have overlapping and unique expression patterns and the 2 genes certainly have overlapping functions (1). However, when combined with pin1–5, yuc4 pin1–5 double mutants displayed a much stronger phenotype than yuc1 pin1–5, indicating that YUC4 plays a more prominent role in inflorescence development than YUC1 (2). It will be interesting to investigate whether other NPY genes can also enhance yuc1 yuc4 and pid.
NPY2 and NPY4 were expressed in Arabidopsis (Fig. 2), but so far we have not had direct evidence that the 2 genes are also involved in auxin-regulated plant development. It is striking that NPY2 was specifically expressed only in the root meristem (Fig. 2A), indicating that NPY2 may have a root-specific function. As discussed previously, npy1 pid double mutants had severe defects in cotyledon development; however, npy1 npy2 npy3 npy4 npy5 quintuple mutants did not display obvious defects in embryogenesis, suggesting that additional NPY genes may also participate in embryogenesis. The obvious candidates would be the 3 genes that have the highest homology to the NPY clade (Fig. 1A). Furthermore, vascular development in cotyledons was disrupted when At5g10250, a distant homolog of NPY1, was mutated, suggesting that more NPH3/NPY-like genes may be involved in auxin-mediated plant development (26).
Control of Cotyledon Development by AGC Kinases.
PID is known mainly for its role in the formation of flowers (16, 17). The fact that simultaneous inactivation of PID and its 3 closest homologs in Arabidopsis abolished the formation of cotyledons (Fig. 4D) demonstrates that the PID genes are also essential for cotyledon development. Remarkably, pid pid2 wag1 wag2 is phenotypically indistinguishable from npy1 pid and yuc1 yuc4 pid (Fig. 4D). Because PID is not a homolog of NPY1 and YUC genes, the genetic enhancement of pid by npy1 or yuc1 yuc4 cannot be explained by gene redundancy. Therefore, we conclude YUC genes, PID, and NPY genes probably are key components in the same pathway involved in regulating organogenesis.
Interestingly, the pid pid2 wag1 wag2 appear to be able to develop a primary root (Fig. 4D), whereas yuc1 yuc4 yuc10 yuc11 failed to make a root meristem during embryogenesis. Therefore, it is likely that additional AGC kinases may be required for the root development (Fig. 1B). It will require us to make the right combinations of AGC kinases to observe defects in other processes.
A Pathway for Auxin-Regulated Organogenesis.
Because YUCs, NPYs, and PIDs all belong to gene families, it is difficult to genetically determine whether they are in the same pathway. It is also difficult to conduct epistatic studies to determine the relative positions of the genes in a pathway. It is clear that members in each of the 3 families have overlapping functions. Plants with multiple YUC genes inactivated have much stronger phenotypes than the single mutants (1, 2). The same is true for the PID family and NPY family (Figs. 3 and 4). If the 3 families participate in the same pathway and are not involved in other pathways, we expect that removal of all YUC genes should lead to the same phenotypes as those caused by disruption of all NPY genes or PID genes. Although mutants without any YUC or NPY or PID activities are not available, our analysis on yuc, pid, and npy mutant combinations and the genetic interactions among the 3 groups of mutants indicate that the 3 family genes are key components of a pathway that regulates auxin-mediated organogenesis. First, pid and npy1 npy5 displayed very similar defects in flower development: both formed pin-like inflorescences and abnormal flowers (Fig. 3). The defects in gynoecium of npy1 npy5 were also similar to those in pid and yuc1 yuc4 quadruple mutants (1), demonstrating that mutations in each gene family lead to similar developmental defects. Second, the formation of cotyledons was completely disrupted by simultaneously inactivating PID, PID2, WAG1, and WAG2. The same cotyledon defects can also be achieved by disrupting both PID and NPY1 or inactivating YUC1, YUC4, and PID simultaneously. We propose that the 3 gene families participate in a linear pathway that regulates auxin-mediated organogenesis. PID and its 3 closest homologs are required for the formation of cotyledons. PID appears to be the most prominent member in the family because pid alone has flower defects and inactivating pid2, wag1, and wag2 does not result in defects in flower development. NPY1 and its homologs NPY3 and NPY5 are required for organogenesis (Fig. 3). Simultaneously inactivating PID and NPY1 would effectively cut the output of the pathway to the level of multiple pid mutants. For example, if PID accounts for 70% of the output of the PID family and NPY1 accounts for 70% of the output of the NPY family, the npy1 pid double mutants will cut down the total output of the pathway to 9% if the 2 families work in the same pathway. The total output of the pathway in pid mutant background can also be further decreased if additional PID homologs are inactivated, which explains why npy1 pid and pid pid2 wag1 wag3 displayed identical cotyledon phenotypes. The same interpretation can also be applied to the synergistic genetic interactions between yuc1 yuc4 and pid.
Our data support that YUCs, PIDs, and NPYs are components of a linear pathway that controls auxin-mediated organogenesis. However, the relative positions of the 3 gene families in the pathway have not been determined. Because PID/NPY-mediated organogenesis is analogous to PHOT1/NPH3-mediated phototropic responses where PHOT1 is upstream of NPH3, we propose that PID is probably upstream of NPY1. Interestingly, NPH3 physically interacts with the chromophore-binding portion of PHOT1 to form a protein complex (21, 27). Unlike PHOT1, PID and its close homologs do not have the photoreceptor domain, suggesting that additional adaptor proteins may be involved in the PID/NPY pathway. Consistent with this hypothesis, preliminary studies indicate that PID and NPY1 do not interact directly (19).
This work has put the complex genetic interactions among YUCs, PIDs, and NPYs into a simple pathway that provides a genetic basis for further understanding of how auxin regulates plant organogenesis. For example, it now becomes feasible to determine the relative positions of the components by gain-of-function studies in the various mutant backgrounds. Our genetic analysis on the gene families shown here also provides a model for analyzing complex genetic interactions among multiple gene families in other pathways.
Methods
Plant Materials.
The T-DNA insertion mutants were obtained from the Arabidopsis Biological Resource Center or The Nottingham Arabidopsis Stock Center (NASC). The npy1 mutant used in this study was the npy1–2 T-DNA allele (SALK-108406), which has been described (18). The npy2 mutant was the T-DNA line SALK-142094, which contained a T-DNA insertion in the first intron of the gene 75 bp after the ATG start codon. The npy3 mutant referred to the T-DNA line SALK-119048. The T-DNA was inserted in the second exon of NPY3, 876 bp downstream of the ATG start codon. The npy4 mutant was the SALK-046452 line, in which the T-DNA was inserted in the fourth exon, 1,628 bp downstream of the ATG start codon of NPY4. The npy5 mutant was the T-DNA line N372878, a GABI-Kat line (GK-027H10) from NASC. The T-DNA was inserted in the fourth exon, 1,746 bp after the start codon of NPY5.
The T-DNA mutants were genotyped by using 3 primers according to the protocol described (28). For npy1, npy2, npy3, and npy4, the mutants were genotyped by using 2 gene-specific primers and 1 T-DNA-specific primer JMLB1. The T-DNA-specific primer for genotyping the T-DNA insertion site in npy5 was PAC106-T-DNA (5′-ATATTGACCATCATACTCATTGC-3′). The gene-specific primers for genotyping npy mutants are shown in Table S1.
The mutants for pid and its homologs all were T-DNA lines. The pid allele was the SALK-049736 line as reported (18). The pid2 allele referred to the line SAIL-269-G07, in which a T-DNA fragment was inserted at 2,010 bp downstream of the ATG start codon of PID2. The wag1 mutant referred to the SALK-002056. The T-DNA insertion was at 372 bp after the ATG start codon of WAG1. The T-DNA allele of wag2 was the line SALK-070240, which had a T-DNA insertion at 207 bp downstream of the ATG start codon of WAG2. The T-DNA-specific primer for genotyping wag1 and wag2 was the JMLB1, and the T-DNA-specific primer for pid2 was SAIL-LB1 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′). The gene specific primers for genotyping the PID homolog mutants are listed in Table S1.
Methods.
We conducted the in situ RNA hybridization according to the methods described (29). The full-length cDNA of NPY and PID genes were used as probes for the in situ hybridization. For in situ hybridization, at least 10 embryos with similar expression patterns were analyzed. Vascular visualization was performed by using protocols from ref. 1.
Supplementary Material
Acknowledgments.
We thank F. Pierri, S. Lee, A. Fang, Y. Liu, Y. Sun, M. Lewis, C. Won, and D. Ashak for DNA preparations and genotyping of various mutants and J. Du for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant R01GM68631 (to Y.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809761106/DCSupplemental.
References
- 1.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Friml J, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 5.Galweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–22230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 6.Aida M, Tasaka M. Genetic control of shoot organ boundaries. Curr Opin Plant Biol. 2006;9:72–77. doi: 10.1016/j.pbi.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Heisler MG, Jonsson H. Modeling meristem development in plants. Curr Opin Plant Biol. 2007;10:92–97. doi: 10.1016/j.pbi.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Nawy T, Lukowitz W, Bayer M. Talk global, act local-patterning the Arabidopsis embryo. Curr Opin Plant Biol. 2008;11:28–33. doi: 10.1016/j.pbi.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Prusinkiewicz P, Rolland-Lagan AG. Modeling plant morphogenesis. Curr Opin Plant Biol. 2006;9:83–88. doi: 10.1016/j.pbi.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith RS, et al. A plausible model of phyllotaxis. Proc Natl Acad Sci USA. 2006;103:1301–1306. doi: 10.1073/pnas.0510457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 13.Hamann T, Mayer U, Jurgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- 14.Przemeck GK, et al. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;200:229–237. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- 15.Hardtke CS, et al. Overlapping and nonredundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- 16.Bennett SRM, Alvarez J, Bossinger G, Smyth DG. Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 1995;8:505–520. [Google Scholar]
- 17.Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100:469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y, Qin G, Dai X, Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furutani M, et al. The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development. 2007;134:3849–3859. doi: 10.1242/dev.009654. [DOI] [PubMed] [Google Scholar]
- 20.Treml BS, et al. The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development. 2005;132:4063–4074. doi: 10.1242/dev.01969. [DOI] [PubMed] [Google Scholar]
- 21.Motchoulski A, Liscum E. Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science. 1999;286:961–964. doi: 10.1126/science.286.5441.961. [DOI] [PubMed] [Google Scholar]
- 22.Bogre L, Okresz L, Henriques R, Anthony RG. Growth signaling pathways in Arabidopsis and the AGC protein kinases. Trends Plants Sci. 2003;8:424–431. doi: 10.1016/S1360-1385(03)00188-2. [DOI] [PubMed] [Google Scholar]
- 23.Harper RM, et al. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galvan-Ampudia CS, Offringa R. Plant evolution: AGC kinases tell the auxin tale. Trends Plants Sci. 2007;12:541–547. doi: 10.1016/j.tplants.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Santner AA, Watson JC. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J. 2006;45:752–764. doi: 10.1111/j.1365-313X.2005.02641.x. [DOI] [PubMed] [Google Scholar]
- 26.Petricka JJ, Clay NK, Nelson TM. Vein patterning screens and the defectively organized tributaries (dot) mutants in Arabidopsis thaliana. Plant J. 2008;56:251–263. doi: 10.1111/j.1365-313X.2008.03595.x. [DOI] [PubMed] [Google Scholar]
- 27.Pedmale UV, Liscum E. Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J Biol Chem. 2007;282:19992–20001. doi: 10.1074/jbc.M702551200. [DOI] [PubMed] [Google Scholar]
- 28.Alonso JM, et al. Genomewide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 29.Dinneny JR, Weigel D, Yanofsky MF. NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development. 2006;133:1645–1655. doi: 10.1242/dev.02335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.