Abstract
Bacterial aromatic polyketides are important therapeutic compounds including front line antibiotics and anticancer drugs. It is one of the last remaining major classes of natural products of which the biosynthesis has not been reconstituted in the genetically superior host Escherichia coli. Here, we demonstrate the engineered biosynthesis of bacterial aromatic polyketides in E. coli by using a dissected and reassembled fungal polyketide synthase (PKS). The minimal PKS of the megasynthase PKS4 from Gibberella fujikuroi was extracted by using two approaches. The first approach yielded a stand-alone Ketosynthase (KS)_malonyl-CoA:ACP transferase (MAT) didomain and an acyl-carrier protein (ACP) domain, whereas the second approach yielded a compact PKS (PKS_WJ) that consists of KS, MAT, and ACP on a single polypeptide. Both minimal PKSs produced nonfungal polyketides cyclized via different regioselectivity, whereas the fungal-specific C2-C7 cyclization mode was not observed. The kinetic properties of the two minimal PKSs were characterized to confirm both PKSs can synthesize polyketides with similar efficiency as the parent PKS4 megasynthase. Both minimal PKSs interacted effectively with exogenous polyketide cyclases as demonstrated by the synthesis of predominantly PK8 3 or NonaSEK4 6 in the presence of a C9-C14 or a C7-C12 cyclase, respectively. When PKS_WJ and downstream tailoring enzymes were expressed in E. coli, the expected nonaketide anthraquinone SEK26 was recovered in good titer. High-cell density fermentation was performed to demonstrate the scale-up potential of the in vivo platform for the biosynthesis of bacterial polyketides. Using engineered fungal PKSs can therefore be a general approach toward the heterologous biosynthesis of bacterial aromatic polyketides in E. coli.
Keywords: cyclase, ketosynthase, megasynthase
Escherichia coli is a powerful microorganism for understanding and engineering the biosynthesis of natural products. This is attributed to its faster growth characteristics, more abundant genetic tools, and a better understanding of its primary metabolism compared with native hosts. Nearly all major classes of natural products have been synthesized and engineered in E. coli, including macrolides (1), cyclic peptides (2), terpenes (3), and alkaloids (4). A noticeable exception has been the pharmaceutically important bacterial aromatic polyketides, which include important human therapeutics such as tetracyclines and anthracyclines. Bacterial aromatic polyketides are synthesized by type II aromatic polyketide synthases (PKSs) from soil-borne actinomycetes (5). The barrier to reconstituting the biosynthesis of aromatic polyketides in E. coli has been the inability to generate the elongated poly-β-ketone backbone from malonyl-CoA, which requires a minimal PKS (6) that consists of a ketosynthase (KS)-chain length factor (CLF) (also known as KSα-KSβ) heterodimer and an acyl-carrier protein (ACP) (Fig. 1B). Despite repeated attempts by numerous groups, expression of the core KS-CLF heterodimer in E. coli has always resulted in 100% of the proteins as inclusion bodies. Therefore, an alternative minimal PKS machinery capable of generating an elongated polyketide backbone, as well as interacting with the immediate tailoring enzymes, is needed to synthesize bacterial aromatic polyketides in E. coli.
Fig. 1.
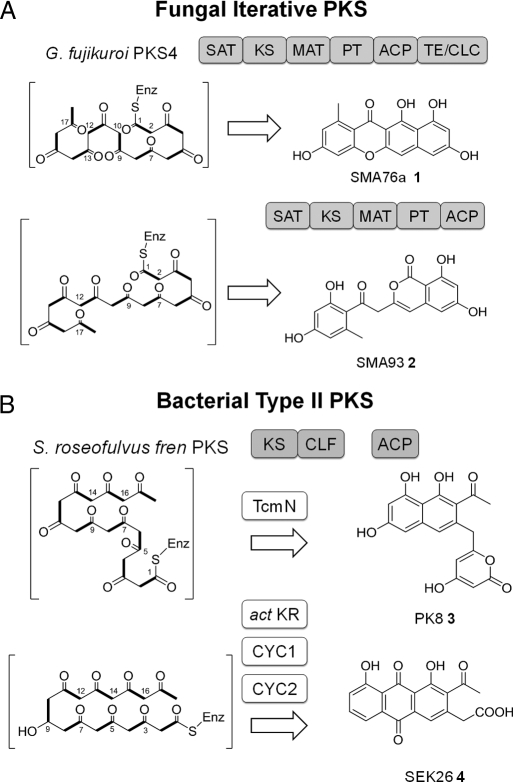
Comparison between fungal and bacterial iterative polyketide synthases. (A) Fungal iterative PKS (IPKS) is a megasynthase that contains all of the catalytic domains on a single polypeptide. The G. fujikuroi PKS4 synthesizes the nonaketide backbone and cyclizes through C2-C7 regioselectivity. The TE/CLC domain catalyzes the C1-C10 cyclization and leads to formation of 1. Without the TE/CLC domain, SMA93 2 is isolated. (B) Bacterial type II PKSs. The minimal PKS consists of the KS-CLF heterodimer and a dissociated ACP domain. The fren minimal PKS is also a nonaketide synthase. However, cyclization of the backbone is determined by dissociated cyclases. The TcmN cyclase can fix the C9-C14 connectivity to form PK8 3. When a defined set of tailoring enzymes is included with the fren minimal PKS, the anthraquinone compound SEK26 4 is formed via reduction of C9 and sequential cyclization of C7-C12, C5-C14, and C3-C16.
The fungal nonreducing PKSs are involved in the biosynthesis of fungal aromatic metabolites, including the well-known mycotoxin aflatoxin (7). Unlike bacterial aromatic PKSs in which the enzymatic components are dissociated, a fungal PKS is a megasynthase that contains the required catalytic domains in a single polypeptide (8) (Fig. 1A). These domains include the starter-unit:ACP acyltransferase (SAT) involved in starter unit selection (9), KS, malonyl-CoA:ACP transferase (MAT) for extender unit loading (10), product-template (PT) (11), ACP, and thioesterase/claisen-cyclase (TE/CLC) (12). The juxtaposition of these domains enables the megasynthase to initiate, extend, and cyclize the polyketide backbone into multicyclic, fungal-specific products (8). We have recently demonstrated that full-length fungal megasynthases can be expressed and reconstituted in E. coli, hence making this family an attractive machinery for E. coli-based biosynthesis of bacterial aromatic polyketides (13). However, despite the analogous iterative Claisen-like condensations catalyzed by two families of PKSs during chain elongation (10), cyclization modes of the poly-β-ketone backbones are drastically different (14, 15) (Fig. 1). For example, both the fungal PKS4 (16) from Gibberella fujikuroi and the bacterial frenolicin (fren) PKS (17) from Streptomyces roseofulvus can synthesize the nonaketide (C18) backbone from 9 malonyl-CoA extender units. The differences in cyclization modes between the two PKSs, however, give rise to orthogonal sets of aromatic polyketide products. PKS4 regioselectively directs the 4 consecutive cyclization reactions, starting with the C2-C7 aldol condensation, to form the tetracyclic SMA76a 1 (13). In the absence of the TE/CLC domain, which is responsible for the second ring cyclization in 1, the C2-C7 cyclized polyketide intermediate spontaneously rearranges to yield the benzopyrone SMA93 2 (18). In contrast, the fren PKS can be combined with an accessory cyclase such as TcmN from the tetracenomycin pathway (19) to cyclize via C9-C14 regioselectivity and afford PK8 3 (20). Similarly, the fren PKS can also interact with a series of tailoring enzymes to yield the anthraquinone SEK26 4 (21).
In this report, we demonstrate the dissection and reassembly of G. fujikuroi PKS4 into a synthetic PKS that can efficiently synthesize bacterial aromatic polyketides. The engineered PKS afforded a spectrum of polyketides with cyclization regioselectivity not observed among fungal polyketides. When the PKS machinery was expressed in E. coli with bacterial tailoring enzymes, complex bacterial aromatic polyketides were produced by this host. Our approach overcomes the barrier of reconstituting bacterial minimal PKS in E. coli, and paves the way for the characterization and engineering of the biosynthesis of type II aromatic polyketides in this powerful organism.
Results and Discussion
Design and Expression of the Fungal Minimal PKS.
We previously demonstrated that bacterial accessory enzymes can interface, albeit with low efficiency, with the PKS4 megasynthase in vitro (18). In those experiments, the major product remained 1 because of the robust, built-in C2-C7 cyclization catalyzed by PKS4. When we coexpressed PKS4 and bacterial cyclases in E. coli, we were only able to isolate 1. This is most likely caused by the inability of the dissociated cyclases to access the poly-β-ketone backbone in vivo.
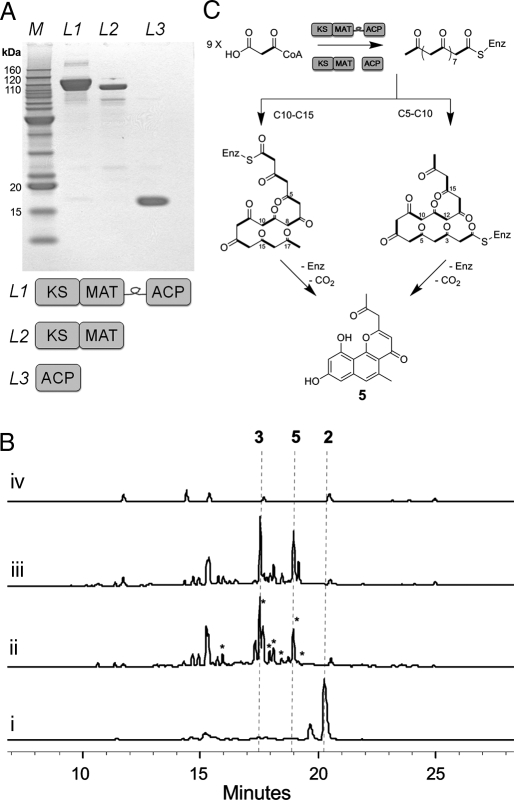
We reasoned the PT domain within PKS4 may be controlling the C2-C7 cyclization. This is supported by the findings of Crawford et al. (7), who showed the PT domain in PksA mediates the consecutive C4-C9, C2-C11 cyclizations required for the synthesis of norsorlinic acid. We devised two strategies to remove the PKS4 PT domain and inactivate the built-in PKS4 cyclization activities (Fig. 2A). In the first approach, the PKS4 KS_MAT didomain and the ACP domain were extracted as stand-alone proteins based on conserved domain boundaries (22). This was designed to mimic the dissociated components of bacterial type II minimal PKS. The KS_MAT (108 kDa) was expressed in E. coli strain BL21(DE3) and the ACP (13 kDa) was expressed in BAP1, an engineered strain of BL21(DE3) that contains a chromosomal copy of the phosphopantetheinyl transferase Sfp (1) (Fig. 2A). In the second approach, both the N terminus SAT and the C terminus TE domains were removed from PKS4, and the entire PT domain was replaced with a KR_ACP linker peptide (≈90 aa) from the 6-deoxyerythronolide synthase (6-DEBS) module 3 (supporting information (SI) Fig. S1 and Tables S1 and S2). The linker was chosen to provide flexibility to the fused ACP domain, allowing it to access the active sites of both KS and MAT domains in cis (22). The design yielded a compact (129 kDa), synthetic megasynthase PKS_WJ that retained all of the minimal PKS components on a single polypeptide. PKS_WJ was solubly expressed in BAP1 with N-terminal hexahistidine tag and was purified to near homogeneity at a final yield of 1.6 mg/L (Fig. 2A).
Fig. 2.
Expression and activities of PKS4 minimal PKS. (A) Construction of PKS4 minimal PKS domains without the PT domain. All proteins were expressed and purified from E. coli with N-terminal 6xHis tags. L1, PKS_WJ (129 kDa); L2, PKS4 KS_MAT didomain (108 kDa); and L3, PKS4 ACP domain (13 kDa). (B) HPLC analysis (280 nm) of minimal PKS activities. All assays used MatB to generate malonyl-CoA in situ. (i) TE-less PKS4 (10 μM) produced 2 as the predominant product. (ii) The dissociated PKS4 minimal PKS (10 μM KS_MAT and 50 μM ACP) synthesized new nonaketides marked with *, while 2 was no longer formed. The major products were 3 and 5. (iii) PKS_WJ (10 μM) produced the similar set of compounds as the dissociated minimal PKS. (iv) No polyketides were produced by the KS_MAT didomain alone (negative control). (C) Two possible cyclization schemes to afford the naphthopyrone 5.
Polyketide Synthesized by the Fungal Minimal PKS.
The catalytic properties of the 2 fungal minimal PKS machineries were examined in the presence of malonate, CoA, and the Rhizobium trifolii malonyl-CoA synthase MatB. Whereas the TE-less PKS4 synthesized predominantly 2, both the dissociated minimal PKS and PKS_WJ produced a similar set of new compounds not observed with the TE-less PKS4 (Fig. 2B). Selected LC/MS ion monitoring of the reaction mixture by using either 12C-malonate or [2-13C]malonate as the malonyl-CoA precursor unambiguously confirmed that many of the new compound (labeled with * in Fig. 2Bii) are derived from nonaketide backbones (Fig. S2). Several unidentified octaketides were also synthesized at trace levels as revealed by MS analysis. Intriguingly, the C2-C7-cyclized product 2 is no longer synthesized by either minimal PKS, indicating that the PT domain is essential for directing the committed cyclization step in the parent megasynthase. As a negative control, removing the ACP component from the dissociated minimal PKS resulted in the abolishment of all polyketide synthesis (Fig. 2Biv), confirming the ACP-dependence of the fungal KS domain.
To obtain more insights into the cyclization patterns of the polyketides synthesized by the fungal minimal PKS, the in vitro reaction was scaled up and the two major products were characterized by 1- and 2-dimensional NMR. Compared with the authentic standard PK8 (Fig. S3), the first major product (RT = 17.4 min) was identified to be the C9-C14-cyclized nonaketide 3, which was previously obtained from an engineered Streptomyces coelicolor strain expressing fren minimal PKS and the C9-C14 cyclase TcmN (20). The second major product (RT = 18.9 min) has parent ion peak [M + H]+ at m/z 299, which is shifted to m/z 308 on the use of [2-13C]malonate as precursor. The mass is in agreement with that of a nonaketide that has been subjected to 3 dehydrations and 1 decarboxylation. Proton NMR revealed the presence of 2 methyl signals and the absence of the commonly observed α-pyrone as a result of O1-C5 esterification, both of which are consistent with the loss of CO2 at C1 of the polyketide. Based on additional NMR evidence and comparison to an authentic standard (Fig. S4), we determined the compound to be the naphthopyrone 5 (Fig. 2C), which was previously isolated as a spontaneously cyclized nonaketide produced by a mutant type III PKS (23). Two possible cyclization modes can lead to the formation of 5 (Fig. 2C). The first intramolecular aldol condensation of the nascent polyketide can take place either between C10 and C15 or C5 and C10, followed by additional cyclization steps, and decarboxylation to yield 5. The minor nonaketide products were not characterized, and were presumably derived from other cyclization regioselectivities. It is surprising that the C7-C12 cyclization regioselectivity commonly observed for type II aromatic polyketides was not found among the major products. Together, the synthesis of 3, 5, and other nonaketides showed that both minimal PKS designs can afford functional enzymes capable of generating a full-length nonaketide backbone, and the committed cyclization modes are no longer C2-C7 as found in products synthesized by parent PKS4 megasynthases.
Kinetic Properties of the Fungal Minimal PKS.
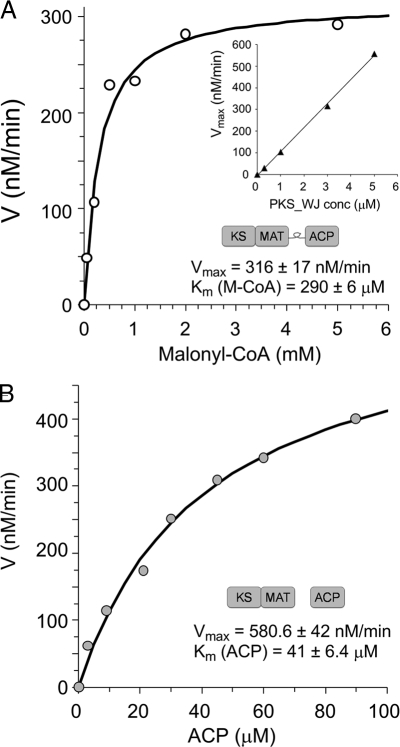
To assess whether the extracted and reconstituted PKS4 minimal PKSs are sufficiently fast to turn over products under in vivo conditions, we quantified the kinetic efficiency of PKS_WJ by using [2-14C]malonyl-CoA and radio-TLC. At 3 μM enzyme concentration, PKS_WJ synthesized polyketides with Vmax = 316 nM/min and displayed Km = 290 μM toward malonyl-CoA with (Fig. 3A). In addition, the maximum velocity of the reaction varied linearly with PKS_WJ concentration to give a kcat = 0.11 min−1 (Fig. 3A Inset). The catalytic efficiency of PKS_WJ is therefore nearly identical to that of the intact PKS4 (kcat = 0.1 min−1) (13). These results provide two important insights into the properties of the synthetic PKS_WJ: (i) removing the SAT and PT domains from the parent PKS4 does not affect the turnover rate of the minimal PKS; (ii) the engineered linker between KS_MAT and ACP in PKS_WJ does not disrupt either KS:ACP or MAT:ACP interaction. The synthetic PKS machinery is therefore kinetically competent to function in E. coli.
Fig. 3.
Kinetic analysis of PKS4 minimal PKS toward nonaketide formation. (A) PKS_WJ (3 μM) exhibited comparable kinetic properties as the parent PKS4, with Vmax = 316 nM/min and Km = 290 μM toward malonyl-CoA. (Inset) The linear correlation between Vmax and PKS_WJ concentration. (B) Michaelis–Menten properties of the dissociated KS_MAT didomain (3 μM) toward the ACP domain at 2 mM malonyl-CoA. The dissociated PKS required a high concentration of ACP (Km = 41 μM) to achieve comparable velocity as PKS_WJ. Data were the average of 3 independent experiments.
We then investigated the kinetic properties of the dissociated minimal PKS. Titration of ACP at 3 μM KS_MAT and 2 mM [2-14C]malonyl-CoA revealed Michaelis–Menten kinetics with Vmax = 581 nM/min and Km (ACP) = 41 μM (Fig. 3B). The high Km requirement to reach a comparable velocity as PKS4 or PKS_WJ is indicative of the catalytic mode in which ACP diffusion and the in trans protein–protein interactions play important roles during catalysis (6), and hints at potential limitations of the dissociated minimal PKS to function efficiently in E. coli. To determine the rate of the malonyl-CoA:ACP transacylation reaction catalyzed by the MAT domain alone, a KS0_MAT mutant (C514A, 10 nM) was used and the rate of [2-14C]malonyl transfer to PKS4 ACP (100 μM) was measured by autoradiography. The apparent rate of malonyl transfer was ≈130 min−1 and is comparable to that of the same reaction catalyzed by freestanding acyltransferases such as S. coelicolor FabD, which is a commonly used MAT in type II PKS assays. Therefore, the rate-limiting step of the minimal PKS is likely one of the catalytic reactions involving the KS domain, including initiation or the Claisen-like condensation extension steps (6).
The in trans protein–protein interactions of the dissociated PKS4 minimal PKS also enabled us to investigate the ACP specificity of the KS_MAT didomain. When PKS4 ACP was substituted with bacterial type I PKS ACPs (DEBS M3 ACP and DEBS M6 ACP), bacterial type II PKS ACPs (DpsG and ZhuN), or heterologous fungal PKS ACPs [LNKS ACP (24) and Gibberella zeae PKS13 ACP (25)], no trace of any polyketides products can be detected by using radio-TLC or LC-MS. This indicates that the interactions between fungal KS_MAT and its ACP is highly specific. By using the PKS4 KS0_MAT mutant didomain, we observed that the MAT domain was able to transfer [2-14C]malonyl label to the heterologous ACP partners with nearly equal efficiency as toward PKS4 ACP. This suggested that the incompatibilities between the PKS4 KS_MAT and heterologous ACPs are entirely attributed to the highly specific KS:ACP interactions. This is in sharp contrast to the tolerance exhibited by the bacterial minimal PKS toward noncognate ACPs, including those extracted from fungal PKSs (10, 24).
Interaction Between PKS4 Minimal PKS and Bacterial Cyclases.
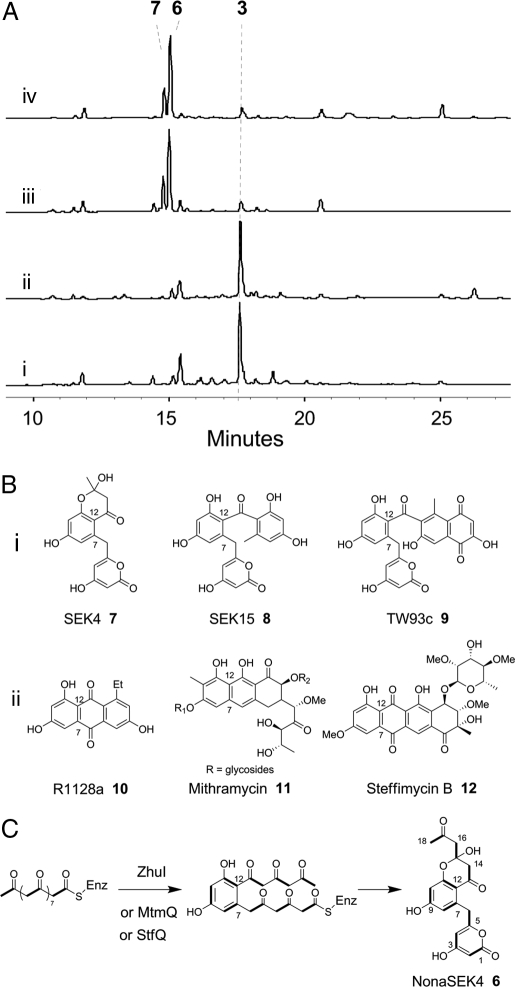
With the minimal PKS fully functional and the built-in C2-C7 cyclization mechanism disabled, we assayed whether cyclization fate of the backbone can now be fully controlled by exogenous cyclases. Remarkably, when the C9-C14 cyclase TcmN or WhiE-ORFVI (26) was added to either minimal PKS, the exclusive synthesis of 3 was observed (Fig. 4Ai and Aii). In both cases, the synthesis of 3 was accompanied by the near total attenuation of other nonaketide products shown in Fig. 2B, illustrating both TcmN and WhiE can completely fix the C9-C14 regioselectivity of the first intramolecular aldol condensation.
Fig. 4.
Control of polyketide cyclization with bacterial cyclases. (A) HPLC analysis (280 nm) of reaction mixtures. Addition of the C9-C14-specific TcmN (i) or WhiE-ORFVI (ii) to PKS_WJ resulted in the exclusive synthesis of 3; whereas addition of the putative C7-C12 cyclases ZhuI (40 μM) (iii) or MtmQ (iv) to PKS_WJ resulted in the synthesis of nonaketide 6 and the minor, octaketide 7. (B) (i) C7-C12 polyketides produced by bacterial minimal PKSs alone without aid of cyclases. SEK4, SEK15, and TW93c are produced by the act, tcm, and whiE minimal PKSs, respectively. (ii) Natural aromatic polyketides with C7-C12-cyclized first ring. (C) Cyclization of the nonaketide backbone to yield 6, which confirms the activities of ZhuI, MtmQ, and StfQ as C7-C12 cyclases specific for unreduced polyketides. See SI Text for MS data and compound characterization.
The engineered fungal minimal PKS presents a unique opportunity to examine an interesting biochemical question regarding type II aromatic PKSs: are there any C7-C12 cyclases that are specific for unreduced polyketide backbones? This question stems from the observation that bacterial minimal PKSs predominantly produce C7-C12 cyclized polyketides (27), including octaketide SEK4 7 (28), decaketide SEK15 8, and dodecaketide TW93c 9 (26). Thus, the C7-C12 cyclization appears to be controlled by the KS-CLFs and no additional cyclases are required. From X-ray crystal structure of act KS-CLF, Keatinge-Clay et al. (29) proposed that the polyketide backbone is buckled at C9 and C10 in the active site of the KS, hence positioning C7 and C12 in close proximity and initiating the aldol condensation. However, putative first-ring cyclases can be found in all gene clusters of aromatic polyketides that cyclize via C7-C12 (Fig. 4B), including that of R1128 10 (ZhuI) (30), mithramycin 11 (MtmQ) (31), and steffimycin 12 (StfQ) (32). The roles of these enzymes have not been clarified because of the lack of a minimal PKS that can facilitate independent assay of C7-C12 cyclase activities.
The PKS4 minimal PKS does not produce any appreciable amounts of C7-C12-cyclized nonaketides and is therefore an ideal enzyme for examining the roles of the putative cyclases. ZhuI, MtmQ, and StfQ were expressed from BL21(DE3) (Fig. S5) and were each added to PKS_WJ or the dissociated minimal PKS. HPLC analysis showed, for each reaction, that the enzymatic mixture produced predominantly a previously unknown nonaketide NonaSEK4 6 with the concomitant disappearance of other nonaketides (Fig. 4 Aiii and Aiv) (Fig. S6). The C7-C12-cyclized octaketide 7 was also synthesized at low levels (Fig. 4 A and Fig. S7). Proton and carbon NMR characterization (Table S3) of 6 revealed the compound is highly similar to the C7-C12-cyclized octaketide 7, with several new signals including the distinguishing aliphatic ketone (δC = 208 ppm). Additional 2-dimensional NMR characterization confirmed the structure of 6 as shown in Fig. 4C, which is a derivative of 7 that contains an extra ketide unit at C16. This molecule is cyclized spontaneously after the regioselective C7-C12 cyclization, which unambiguously verifies the functions of the 3 cyclases. Their roles in the respective pathways may be to ensure complete C7-C12 cyclization, as well as an increase in the rate of the committed cyclization step. Compound 6 is not synthesized by fungal minimal PKSs, which further confirms the inability of the PKS4 minimal PKS to direct the C7-C12 cyclization and points to a different orientation of the growing polyketide chain in the KS active site compared with that of bacterial KS-CLF. The synthesis of 6 suggests that, although the fungal minimal PKS may have some template influence over the orientation of the nascent polyketide chain to afford 3 and 5 as the major products, addition of polyketide cyclases can completely reorient the polyketide backbone and lead to the formation of highly specific products.
Biosynthesis of Aromatic Polyketides in E. coli.
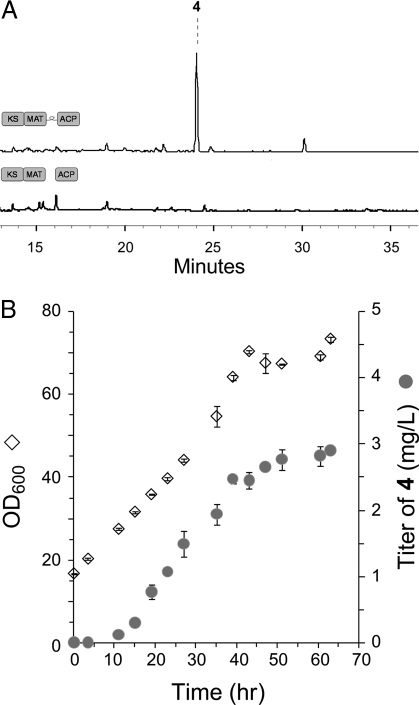
The in vitro data presented so far provide convincing evidence that a fungal minimal PKS can be a functional alternative to a type II minimal PKS. To demonstrate the application of this new enzymatic machinery in E. coli, we targeted the biosynthesis of anthraquinone 4. Compound 4 is derived from a nonaketide backbone after ketoreduction at C9 by a ketoreductase (KR), C7-C12 cyclization/dehydration by an aromatase/cyclase (CYC1), C5-C14 cyclization by a second-ring cyclase (CYC2), followed by spontaneous third-ring cyclization and second-ring oxidation (21). Two plasmids encoding PKS_WJ, act KR, gris CYC1, and oxy CYC2, with each gene flanked by a T7 promoter and a T7 terminator, were transformed into BAP1. The resulting strain was grown in shake flask culture, whereas protein expression was induced with IPTG and continued at 16 °C for 2 days. The expression of all 4 PKS enzymes was monitored by SDS/PAGE, and maximum soluble protein expression levels were reached 24 h after induction. Gratifyingly, analysis of the culture extract by HPLC and LC-MS revealed 4 was indeed synthesized by our engineered E. coli strain at ≈0.4 mg/L (Fig. 5A). The retention time, UV spectrum, and mass patterns all matched precisely to those of SEK26 purified from CH999/pSEK26 (Fig. S8). ≈90% of 4 were found in the culture media, whereas the remaining 10% were extracted from the cell pellets. Selected ion monitoring of the extract showed that no other nonaketide species were present, confirming the precise timing and specificities of the three tailoring enzymes in E. coli. Furthermore, to explore the scale-up potential of this process, we performed a fed-batch fermentation by using F1 minimal medium and monitored the level of 4 for ≈60 h after induction (Fig. 5B). The E. coli culture started producing detectable amounts of 4 ≈10 h after induction, and the titer increased steadily until a maximum of ≈3 mg/L was achieved at a plateau OD600 of ≈70. Prolonged fermentation beyond the plateau did not result in an increase in polyketide levels, which may be attributed to loss of enzyme activity or depleting intracellular malonyl-CoA levels.
Fig. 5.
Biosynthesis of bacterial aromatic polyketides in E. coli. (A) HPLC analysis (430 nm) of SEK26 4 biosynthesis by using E. coli BAP1 expressing PKS_WJ and three tailoring enzymes act KR, gris ARO/CYC, and OxyN. The titer of 4 in shake flask fermentation was ≈0.4 mg/L. (B) High-cell density, fed-batch fermentation production of 4. The maximum titer achieved was ≈3 mg/L at OD600 of ≈70, 60 h after IPTG addition.
In contrast to PKS_WJ, no polyketides can be detected when the dissociated PKS4 minimal PKS was expressed in E. coli despite repeated optimization attempts (Fig. 5A). SDS/PAGE analysis confirmed that both proteins were expressed in the BAP1 host (Fig. S5). The lack of product turnover is possibly due to the high Km of ACP toward the stand-alone KS_MAT enzyme as shown in the kinetic analysis (Fig. 3B). The less efficient protein–protein interactions between dissociated components may not be able to sustain the synthesis of the elongated poly-β-ketone backbone under in vivo conditions. Therefore, although both designs of minimal PKS were catalytically competent in vitro, the synthetic linker we introduced to religate the KS_MAT and ACP domains proved to be essential in facilitating polyketide synthesis in E. coli.
In summary, we have demonstrated the total biosynthesis of bacterial aromatic polyketides in the workhorse organism E. coli. The enabling protein-engineering steps were the extraction and reassembly of the minimal PKS from a nonreducing fungal megasynthase. The extraction step was important to inactivate the built-in cyclization modes of fungal megasynthase, and the reassembly step was indispensable toward establishing a kinetically competent enzyme in E. coli. This approach can be extended to other fungal megasynthases of different chain-length specificities and be applied toward the synthesis of other type II bacterial aromatic polyketides. For example, pharmaceutically relevant decaketides such as anthracyclines can be synthesized in E. coli by reengineering a fungal decaketides synthase from the hypomycetin biosynthetic pathway (33). The genetically superior E. coli system can also provide immense opportunities toward biochemical understanding and protein engineering of aromatic PKS enzymes.
Materials and Methods
Cloning and Expression of the Minimal PKSs.
See SI Text.
Polyketide Product Detection.
Example given for a typical reaction: 10 μM KS_MAT (with 50 μM ACP) or PKS_WJ was incubated with 20 μM MatB, 100 mM sodium malonate (12C- or 2-13C-labeled), 7 mM MgCl2, 5 mM CoA, and 20 mM ATP. The bacterial tailoring enzymes were each added to a final concentration of 40 μM. All polyketide product assays were incubated at room temperature. The assays were stopped after 4 h and extracted 3 times with 99% ethyl acetate (EA)/1% acetic acid (AcOH). The organic phase was separated, evaporated to dryness, redissolved in 20 μl of methanol, and the reaction mixture was subjected to HPLC analysis. HPLC (Alltech Apollo 5u, 250 mm × 4.6 mm) is on a linear gradient of 5–95% CH3CN (vol/vol) >30 min and 95% CH3CN (vol/vol) further for 10 min in H2O supplemented with 0.1% (vol/vol) trifluoroacetic acid at a flow rate of 1.0 ml/min. HPLC retention times were as follows: SMA76a (1), 28.3 min; SMA93 (2), 20.4 min; PK8 (3), 17.4 min; SEK26 (4), 24.0 min; (5), 18.9 min; (6), 15.0 min; (7), 14.7 min.
LC-MS was conducted with a Shimadzu 2010 EV Liquid Chromatography Mass Spectrometer by using both positive and negative electrospray ionization and a Phenomenex Luna 5u 2.0 × 100 mm C18 reverse-phase column. Samples were separated on a linear gradient of 5 to 95% CH3CN (vol/vol) >30 min and 95% CH3CN (vol/vol) further 30 min in H2O supplemented with 0.05% (vol/vol) formic acid at a flow rate of 0.1 ml/min at room temperature.
Polyketide Turnover Assay.
A time course analysis was performed on each of the PKS described in this work. The KS_MAT, PKS_WJ, and PKS4 concentrations were fixed at 3 μM in phosphate buffer (pH 7.4). For malonyl-CoA titration, [2-14C]malonyl-CoA concentrations were varied between 50 μM and 5 mM. For ACP titration, ACP concentrations were varied between 5 and 90 μM and [2-14C]malonyl-CoA concentration was fixed at 2 mM (1.22 nCi/nmol). The reaction was initiated by the addition of [2-14C]malonyl-CoA and allowed to react at room temperature. At each time point (5, 10, and 20 min), a 10-μl aliquot was quenched and extracted with 300 μl of 99% EA/1% AcOH. The organic phase was evaporated to dryness, redissolved in 15 μl EA, separated by TLC (EA/AcOH, 99:1), and quantified with a PhosphorImager.
Small-Scale Shake Flask Low-Density Fermentation.
E. coli BAP1 strain transformed with expression plasmid(s) was grown in F1 medium at 37 °C to an OD600 of 0.4–0.6, at which time the cultures were cooled to 16 °C, and then induced with 0.2 mM IPTG at 250 rpm and grown at 16 °C for 2 days. For product detection, 1 ml of cell culture was collected. The cell pellet was extracted with acetone, evaporated to dryness, and redissolved in methanol. The medium was extracted with 99% EA/1% AcOH, the organic phase was separated, evaporated to dryness, and redissolved in methanol. Both fractions were then subjected to HPLC analysis.
High-Density F1 Fed-Batch Fermentation.
Methods for F1 fed-batch fermentation and medium composition were adopted from methods described by Pfeifer et al. (34). For details, see SI Text.
Supplementary Material
Acknowledgments.
We thank Prof. Ikuro Abe for an authentic standard 5; Prof. Kenji Watanabe for pKW423; Prof. Chaitan Khosla for pPK8, pSEK26, and pSEK4; Prof. Sheryl Tsai for pETWhiE; and Suzanne Ma for providing the purified MatB sample. This was supported by National Institutes of Health Grant 1R01GM085128 and by the David and Lucile Packard Fellowship in Science and Engineering. W. Zhang was supported in part by a Nell I. Mondy Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.H.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809084105/DCSupplemental.
References
- 1.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe K, et al. Total biosynthesis of antitumor nonribosomal peptides in Escherichia coli. Nat Chem Biol. 2006;2:423–428. doi: 10.1038/nchembio803. [DOI] [PubMed] [Google Scholar]
- 3.Chang MC, Eachus RA, Trieu W, Ro DK, Keasling JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol. 2007;3:274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- 4.Minami H, et al. Microbial production of plant benzylisoquinoline alkaloids. Proc Natl Acad Sci USA. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat Prod Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 6.Beltran-Alvarez P, Cox RJ, Crosby J, Simpson TJ. Dissecting the component reactions catalyzed by the actinorhodin minimal polyketide synthase. Biochemistry. 2007;46:14672–14681. doi: 10.1021/bi701784c. [DOI] [PubMed] [Google Scholar]
- 7.Crawford JM, et al. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox RJ. Polyketides, proteins and genes in fungi: Programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 9.Crawford JM, Dancy BC, Hill EA, Udwary DW, Townsend CA. Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc Natl Acad Sci USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, et al. Catalytic relationships between type I and type II iterative polyketide synthases: The Aspergillus parasiticus norsolorinic acid synthase. Chembiochem. 2006;7:1951–1958. doi: 10.1002/cbic.200600341. [DOI] [PubMed] [Google Scholar]
- 11.Udwary DW, Merski M, Townsend CA. A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a type I polyketide synthase. J Mol Biol. 2002;323:585–598. doi: 10.1016/s0022-2836(02)00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem Biol. 2001;8:189–197. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- 13.Ma SM, et al. Enzymatic synthesis of aromatic polyketides using PKS4 from Gibberella fujikuroi. J Am Chem Soc. 2007;129:10642–10643. doi: 10.1021/ja074865p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas R. A biosynthetic classification of fungal and streptomycete fused-ring aromatic polyketides. Chembiochem. 2001;2:612–627. doi: 10.1002/1439-7633(20010903)2:9<612::AID-CBIC612>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 15.Bringmann G, et al. Different polyketide folding modes converge to an identical molecular architecture. Nat Chem Biol. 2006;2:429–433. doi: 10.1038/nchembio805. [DOI] [PubMed] [Google Scholar]
- 16.Linnemannstons P, et al. The polyketide synthase gene pks4 from Gibberella fujikuroi encodes a key enzyme in the biosynthesis of the red pigment bikaverin. Fungal Genet Biol. 2002;37:134–148. doi: 10.1016/s1087-1845(02)00501-7. [DOI] [PubMed] [Google Scholar]
- 17.Bibb MJ, Sherman DH, Omura S, Hopwood DA. Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin. Gene. 1994;142:31–39. doi: 10.1016/0378-1119(94)90351-4. [DOI] [PubMed] [Google Scholar]
- 18.Ma SM, et al. Redirecting the cyclization steps of fungal polyketide synthase. J Am Chem Soc. 2008;130:38–39. doi: 10.1021/ja078091o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen B, Hutchinson CR. Deciphering the mechanism for the assembly of aromatic polyketides by a bacterial polyketide synthase. Proc Natl Acad Sci USA. 1996;93:6600–6604. doi: 10.1073/pnas.93.13.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer PJ, et al. Rational design and engineered biosynthesis of a novel 18-carbon aromatic polyketide. J Am Chem Soc. 1997;119:635–639. [Google Scholar]
- 21.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature. 1995;375:549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci USA. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe I, et al. Structure-based engineering of a plant type III polyketide synthase: Formation of an unnatural nonaketide naphthopyrone. J Am Chem Soc. 2007;129:5976–5980. doi: 10.1021/ja070375l. [DOI] [PubMed] [Google Scholar]
- 24.Ma SM, Tang Y. Biochemical characterization of the minimal polyketide synthase domains in the lovastatin nonaketide synthase LovB. FEBS J. 2007;274:2854–2864. doi: 10.1111/j.1742-4658.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhou H, Zhan J, Watanabe K, Xie X, Tang Y. A polyketide macrolactone synthase from the filamentous fungus Gibberella zeae. Proc Natl Acad Sci USA. 2008;105:6249–6254. doi: 10.1073/pnas.0800657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu TW, et al. Engineered biosnthesis of novel polyketides from streptomyces spore pigment polyketide synthases. J Am Chem Soc. 1998;120:7749–7759. [Google Scholar]
- 27.McDaniel R, Ebert-Khosla S, Fu H, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: Influence of a downstream enzyme on the catalytic specificity of a minimal aromatic polyketide synthase. Proc Natl Acad Sci USA. 1994;91:11542–11546. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu H, Ebertkhosla S, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides : Dissection of the catalytic specificity of the act ketoreductase. J Am Chem Soc. 1994;116:4166–4170. [Google Scholar]
- 29.Keatinge-Clay AT, Maltby DA, Medzihradszky KF, Khosla C, Stroud RM. An antibiotic factory caught in action. Nat Struct Mol Biol. 2004;11:888–893. doi: 10.1038/nsmb808. [DOI] [PubMed] [Google Scholar]
- 30.Marti T, Hu ZH, Pohl NL, Shah AN, Khosla C. Cloning; nucleotide sequence; and heterologous expression of the biosynthetic gene cluster for R1128; a non-steroidal estrogen receptor antagonist : Insights into an unusual priming mechanism. J Biol Chem. 2000;275:33443–33448. doi: 10.1074/jbc.M006766200. [DOI] [PubMed] [Google Scholar]
- 31.Lombo F, Blanco G, Fernandez E, Mendez C, Salas JA. Characterization of Streptomyces argillaceus genes encoding a polyketide synthase involved in the biosynthesis of the antitumor mithramycin. Gene. 1996;172:87–91. doi: 10.1016/0378-1119(96)00029-7. [DOI] [PubMed] [Google Scholar]
- 32.Gullon S, et al. Isolation, characterization, and heterologous expression of the biosynthesis gene cluster for the antitumor anthracycline steffimycin. Appl Environ Microbiol. 2006;72:4172–4183. doi: 10.1128/AEM.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breinholf J, Jensen GW, Kjaer A, Olsen CE, Rosendahl CN. Hypomycetin: an antifungal, tetracycline metabolite from Hypomyces aurantius: Production, structure and biosynthesis. Acta Chem Scandi. 1997;51:855–860. [Google Scholar]
- 34.Pfeifer B, Hu Z, Licari P, Khosla C. Process and metabolic strategies for improved production of Escherichia coli-derived 6-deoxyerythronolide B. Appl Environ Microbiol. 2002;68:3287–3292. doi: 10.1128/AEM.68.7.3287-3292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.