Abstract
Aminoglycoside ototoxicity has been related to a surprisingly large number of cellular structures and metabolic pathways. The finding that patients with mutations in mitochondrial rRNA are hypersusceptible to aminoglycoside-induced hearing loss has indicated a possible role for mitochondrial protein synthesis. To study the molecular interaction of aminoglycosides with eukaryotic ribosomes, we made use of the observation that the drug binding site is a distinct domain defined by the small subunit rRNA, and investigated drug susceptibility of bacterial hybrid ribosomes carrying various alleles of the eukaryotic decoding site. Compared to hybrid ribosomes with the A site of human cytosolic ribosomes, susceptibility of mitochondrial hybrid ribosomes to various aminoglycosides correlated with the relative cochleotoxicity of these drugs. Sequence alterations that correspond to the mitochondrial deafness mutations A1555G and C1494T increased drug-binding and rendered the ribosomal decoding site hypersusceptible to aminoglycoside-induced mistranslation and inhibition of protein synthesis. Our results provide experimental support for aminoglycoside-induced dysfunction of the mitochondrial ribosome. We propose a pathogenic mechanism in which interference of aminoglycosides with mitochondrial protein synthesis exacerbates the drugs' cochlear toxicity, playing a key role in sporadic dose-dependent and genetically inherited, aminoglycoside-induced deafness.
Keywords: decoding, mitochondria, ribosomes, toxicity, translation
Low cost and high efficacy make aminoglycosides a common choice for treatment of serious infections caused by gram-negative bacilli, including endocarditis, sepsis, pneumonia, pyelonephritis, and multidrug-resistant tuberculosis (1). Unfortunately, aminoglycosides are both nephrotoxic and ototoxic. Although renal impairment is in general mild and reversible, ototoxicity results from drug-induced apoptosis of cochlear and vestibular hair cells and is irreversible (2, 3). Ototoxicity of aminoglycoside antibiotics occurs both in a dose-dependent and in an inherited idiosyncratic fashion. Despite attempts to limit drug doses and to monitor blood levels carefully, measurable signs of hearing loss are found in 20% of patients receiving aminoglycosides (2). Familial cases of aminoglycoside-induced deafness are maternally transmitted and linked to mutations in mitochondrial DNA (mtDNA) (4–6).
The mechanisms by which aminoglycoside antibiotics exert their toxic effects are controversial. A surprisingly large and diverse number of effects have been associated with aminoglycosides. Aminoglycosides have been reported to affect DNA, RNA, and protein synthesis; energy metabolism and ion transport; and synthesis or degradation of prostaglandins, gangliosides, mucopolysaccharides and lipids (2). In addition, it has been hypothesized that aminoglycosides may form cochleotoxic metabolites. Antioxidants apparently attenuate aminoglycoside-induced hearing loss, pointing to a role of the mitochondrion, an organelle involved in oxidation, as a target of ototoxic drugs (7, 8). Genetic analyses of individuals hypersensitive to aminoglycosides have identified mutations in mitochondrial rRNA. Transition mutations in the mitochondrial small ribosomal RNA gene, namely A1555G and, less frequently C1494T, have been identified as primary genetic traits in aminoglycoside-induced deafness (4, 6, 9). A1555G and C1494T both map to the aminoacyl-tRNA acceptor site (A site) of the small ribosomal subunit. The bacterial A-site rRNA is target for aminoglycoside antibiotics, which exert their antibacterial effect at the level of the prokaryotic ribosome (10–13). Aminoglycosides affect protein synthesis by inducing codon misreading and by inhibiting translocation of the tRNA-mRNA complex (14, 15). The basis for aminoglycoside selectivity is presumably their preferential binding to the bacterial as opposed to eukaryotic ribosomes (13, 16, 17).
The high copy number of mtDNA in mitochondria and the vast number of mitochondria in a single cell have frustrated any attempt of genetic manipulation of mitochondrial rRNA in lower and higher eukaryotes. Model oligonucleotides designed to mimic the drug-binding site have been used to investigate various aspects of aminoglycoside-ribosome interaction (18–22). However, conclusions derived from the study of model A-site oligonucleotides are compromised by several findings: (i) in contrast to drug susceptibility of complete ribosomes, binding affinities of aminoglycosides to prokaryotic decoding region constructs are not very sensitive to mutations within the RNA-binding region (23); (ii) in vivo drug susceptibilities of mutant ribosomes and in vitro binding affinities using variants of model A-site oligonucleotides may or may not correlate (24–26); (iii) the exquisite specificity of aminoglycosides for the prokaryotic as opposed to the eukaryotic cytosolic ribosome contrasts with the observation that these drugs bind to eukaryotic decoding-site constructs with approximately the same affinity as found for their prokaryotic counterpart (23, 24); and (iv) while there is evidence that mitochondrial ribosomes are susceptible to aminoglycosides (13, 27), oligonucleotides mimicking the mitochondrial A site do not bind aminoglycosides to any significant extent (24, 28).
Using gene-shuffling experiments, we have previously replaced the A-site residues of helix 44 (H44) in bacterial 16S rRNA with various eukaryotic homologues, demonstrating that the A-site rRNA behaves as an autonomous domain, which can be exchanged between different species for study of function (29, 30). Replacement of a 34-nucleotide portion of bacterial 16S-rRNA helix 44 with its human homologues resulted in rRNA-decoding sites virtually identical to that in cytosolic and mitochondrial ribosomes. Here we used hybrid bacterial ribosomes carrying distinct alleles of the mitochondrial decoding site to study aminoglycoside susceptibility of wild-type and mutant mitochondrial rRNA.
Results and Discussion
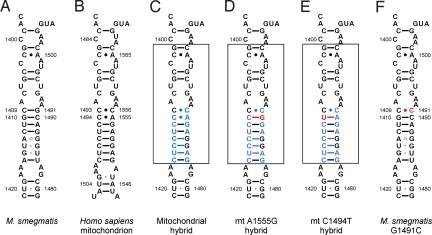
The in vivo activity of various 2-deoxystreptamine antibiotics against isogenic Mycobacterium smegmatis strains carrying mitochondrial-bacterial hybrid ribosomes was tested in minimal inhibitory concentration (MIC) assays, which determine growth inhibition at the whole-cell level. Compared to bacterial ribosomes, which were found to be unanimously susceptible to all aminoglycosides tested, the hybrid ribosomes with a wild-type mitochondrial H44 revealed a heterogeneous drug susceptibility pattern, with MIC values ranging from 32 to 1,024 μg/ml (Table 1). The ratio of MIC mitochondrial hybrid to MIC wild-type M. smegmatis varied from 64-fold (gentamicin, amikacin) to 256-fold (netilmicin, kanamycin), providing a relative measure of the drug-target selectivity of different 2-deoxystreptamine antibiotics. We next investigated recombinants where the bacterial H44 has been replaced by mitochondrial deafness alleles corresponding to mtDNA mutations A1555G and C1494T. The resulting mutant mitochondrial hybrid ribosomes differ from the wild-type mitochondrial hybrid only in 16S rRNA residues 1490 and 1410; compare Fig. 1C to Fig. 1 D and E (bacterial 16S rRNA residues are numbered according to Escherichia coli nomenclature). The presence of the A1555G or the C1494T mutation increased drug susceptibility of cells carrying the mitochondrial hybrid ribosomes by 4- to 16-fold (see Table 1).
Table 1.
Minimal inhibitory concentrations (μg/ml) of aminoglycoside antibiotics
| Aminoglycoside | A-site rRNA |
|||
|---|---|---|---|---|
| Bacteriala | Mitochondrial (mt) hybrid | mt A1555G hybrid | mt C1494T hybrid | |
| Neomycin B | 0.5 | 16–32 | 8 | 8 |
| Paromomycin | 1 | > 1,024 | 256–512 | 256–512 |
| Kanamycin A | 1 | 256–512 | 16–32 | 16 |
| Tobramycin | 1 | 128 | 16 | 16 |
| Amikacin | 0.5 | 32–64 | 2–4 | 2–4 |
| Gentamicin | 1 | 64–128 | 16–32 | 16–32 |
| Netilmicin | 2 | 512–1024 | 64 | 64–128 |
aM. smegmatis wild-type rRNA
Fig. 1.
Secondary structure of rRNA helix 44 in the ribosomal decoding site. (A) Decoding site of M. smegmatis wild-type ribosomes; rRNA nucleotides are numbered according to the bacterial nomenclature (i.e., homologous E. coli 16S rRNA positions). (B) Decoding site of human mitochondrial ribosomes; rRNA residues are numbered according to the mitochondrial nomenclature. (C–E) Mitochondrial decoding sites within human-bacterial hybrid ribosomes: wild type sequence (C) and deafness-associated alterations adenine to guanine at position 1490 (corresponding to mitochondrial mutation A1555G) (D); cytosine to uracil mutation at position 1410 (corresponding to mitochondrial mutation C1494T) (E). Nucleotide positions depicted in blue represent residues that are specific for human rRNA; nucleotide positions in red highlight the pathogenic mutations; the transplanted helix is boxed. (F) Decoding site of the M. smegmatis G1491C mutant.
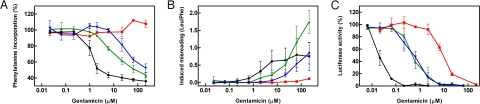
For a more detailed study of the mt A1555G and C1494T alleles, we studied purified hybrid ribosomes in cell-free translation reactions. We first used an AUG(UUU)12-mRNA template, as this message allows determination of drug-induced inhibition of polypeptide synthesis and amino acid misincorporation. Dose-response curves of aminoglycoside-induced inhibition of phenylalanine incorporation were analyzed to define the IC50 values of the individual 2-deoxystreptamines. Both the A1555G and the C1494T genotypes were more susceptible to aminoglycoside antibiotics than the wild-type mitochondrial decoding site, as indicated by the finding that significantly less drug concentrations were required to inhibit AUG(UUU)12 mRNA-driven polyPhe synthesis [see Fig. 2A, Table 2, and supporting information (SI) Fig. S1].
Fig. 2.
Aminoglycoside susceptibility of mutant and wild-type mitochondrial hybrid ribosomes. Dose-response curves of wild-type mitochondrial (red squares), mutant A1555G (green triangles), and mutant C1494T (blue inverted triangles) hybrid ribosomes; bacterial ribosomes (black circles) are included for comparison. (A) Gentamicin-induced inhibition of [14C]-phenylalanine incorporation (n ≥ 3; ± SD.). The 100% value corresponds to 25 to 30 pmol Phe incorporation. Corresponding IC50 values of gentamicin and selected aminoglycoside antibiotics are presented in Table 2. (B) Gentamicin-induced increase in misincorporation of the near-cognate [3H]-leucine relative to the drug-free control (n ≥ 3; ± SD.). (C) Gentamicin-induced inhibition of luciferase synthesis relative to the drug-free control (n ≥ 3; ± SD.). Corresponding IC50 values for gentamicin and selected aminoglycoside antibiotics are presented in Table 3.
Table 2.
Aminoglycoside-induced inhibition of AUG(UUU)12-driven phenylalanine incorporation (IC50, μM)
| Aminoglycoside | A-site rRNA |
|||
|---|---|---|---|---|
| Bacteriala | Mitochondrial (mt) hybrid | mt A1555G hybrid | mt C1494T hybrid | |
| Neomycin B | 0.2 ± 0.0 | 209 ± 61 | 3.1 ± 0.3 | 3.7 ± 0.7 |
| Paromomycin | 0.7 ± 0.1 | > 500 | 124 ± 35 | 116 ± 20 |
| Kanamycin A | 1.2 ± 0.2 | > 500 | 20 ± 2 | 95 ± 29 |
| Tobramycin | 0.7 ± 0.1 | > 500 | 15 ± 3 | 70 ± 23 |
| Amikacin | 0.8 ± 0.1 | > 500 | 7.1 ± 0.8 | 13 ± 3 |
| Gentamicin | 1.3 ± 0.2 | > 500 | 13 ± 2 | 62 ± 16 |
| Netilmicin | 1.2 ± 0.3 | > 500 | 71 ± 10 | 331 ± 84 |
IC50 values represent the drug concentrations in μM that are required to inhibit AUG(UUU)12-driven phenylalanine incorporation to half-maximal extent. A representative graph showing phenylalanine incorporation plotted against gentamicin concentration is shown in Fig. 2A.
aM. smegmatis wild-type rRNA
Aminoglycosides are known to affect the translational fidelity of ribosomes by inducing misreading of the genetic code (14). For study of aminoglycoside-induced mistranslation we used the AUG(UUU)12-driven polypeptide synthesis assay to determine the relative amount of near-cognate leucine incorporation compared to incorporation of the cognate amino acid phenylalanine in the presence of various concentrations of gentamicin. Relative incorporation of [3H]-labeled leucine versus [14C]-labeled phenylalanine was determined and plotted against gentamicin concentration. Introduction of the A1555G and C1494T alteration rendered the mitochondrial hybrid ribosomes highly susceptible to aminoglycoside-induced misreading (Fig. 2B; see also Fig. S2). In quantitative terms, the amount of gentamicin-induced misreading (calculated as leucine per phenylalanine incorporation) in A1555G and C1494T mutant hybrid ribosomes was up to 1.75 leucine per phenylalanine, as compared to a maximum of 0.2 leucine per phenylalanine for hybrid ribosomes with a wild-type mitochondrial decoding site.
To study the effect of aminoglycoside antibiotics on translation of a more natural mRNA template, we tested wild-type and mutant mitochondrial hybrid ribosomes in a cell-free luciferase synthesis assay. As depicted in Fig. 2C, Table 3, and Fig. S3, the allele- and drug-specific inhibition of luciferase synthesis essentially correlated with the results of the MIC and AUG(UUU)12 assays. Drug-mediated inhibition of luciferase synthesis was significantly increased in A1555G and C1494T mutant ribosomes.
Table 3.
Aminoglycoside-induced inhibition of luciferase synthesis (IC50, μM)
| Aminoglycoside | A-site rRNA |
|||
|---|---|---|---|---|
| Bacteriala | Mitochondrial (mt) hybrid | mt A1555G hybrid | mt C1494T hybrid | |
| Neamine | 1.4 | 131 | 32 | 35 |
| Neomycin B | 0.04 | 0.7 | 0.5 | 0.4 |
| Paromomycin | 0.03 | 33 | 2.4 | 3.1 |
| Kanamycin A | 0.05 | 15.7 | 1.2 | 1.1 |
| Tobramycin | 0.02 | 7.8 | 0.8 | 0.9 |
| Amikacin | 0.02 | 7.0 | 0.4 | 0.6 |
| Gentamicin | 0.03 | 5.7 | 0.6 | 0.7 |
| Netilmicin | 0.05 | 17.6 | 0.8 | 2.6 |
IC50 values represent the drug concentrations in μM that are required to inhibit synthesis of functional firefly luciferase to 50%. Relative luciferase activity plotted against aminoglycoside concentration is shown in Fig. 2C.
aM. smegmatis wild-type rRNA
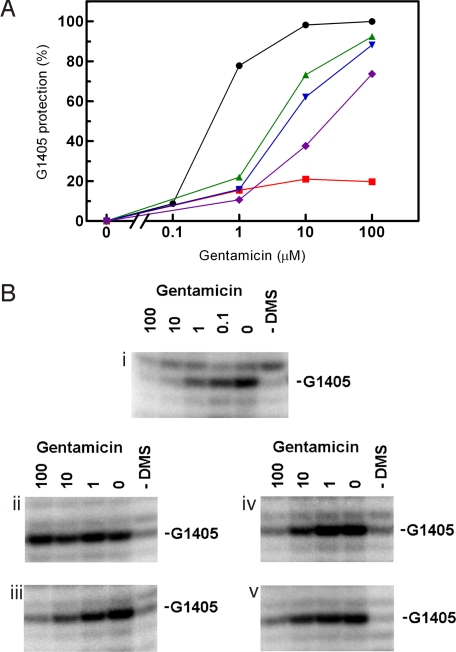
The basis for the selectivity of aminoglycosides is presumably their preferential binding to bacterial as opposed to eukaryotic ribosomes (10, 13, 16, 17). In particular, 16S rRNA nucleotides 1408, 1409, and 1491 of helix 44 have been shown to be critical for drug-binding by forming direct contacts with ring I of the 2-deoxystreptamines (12, 13, 16, 17, 26, 31–38) (see Fig. S4 for the chemical structures of aminoglycosides used in this study). In the absence of X-ray structures for aminoglycosides complexed to the mitochondrial ribosome, we can rationalize our findings on data invoked from the study of bacterial ribosome-drug complexes (12, 39). The rRNA secondary structure of the drug binding site in mitochondrial A1555G and C1494T mutant ribosomes resembles that of bacterial ribosomes with a G1491C alteration in that the C1409-C1491 opposition is accompanied by a 1410–1490 Watson–Crick pair (see Fig. 1). The bacterial G1491C ribosome shows a drug-susceptibility phenotype that is virtually superimposable on that found for the mitochondrial deafness alleles (see Table S1 for comparison). To determine whether the affinity of aminoglycosides to mutant mitochondrial decoding sites corresponds to that of the bacterial G1491C decoding site, we probed gentamicin binding by chemical footprinting experiments. Bacterial wild-type ribosomes showed drug-mediated protection from dimethyl sulfate (DMS) modification at G1405 (N-7), which is in good agreement with previous reports on aminoglycoside protection in bacterial 16S rRNA (34). Wild-type mitochondrial hybrid ribosomes showed little protection, while mutant mitochondrial A1555G and C1494T hybrid ribosomes showed a concentration-dependent protection of G1405 that resembles the dose-response curve observed with bacterial G1491C ribosomes (Fig. 3). Thus, binding of aminoglycosides to ribosomes with an adenine at 16S rRNA position 1408, appears to be mainly determined by the structural geometry of base pairs 1409–1491 and 1410–1490.
Fig. 3.
Chemical footprints of gentamicin binding to wild-type and mutant mitochondrial decoding sites in comparison to bacterial wild-type and G1491C ribosomes. (A) Gentamicin-dependent protection of G1405 in wild-type mitochondrial (red squares), mutant mt A1555G (green triangles), mutant mt C1494T (blue inverted triangles), and bacterial G1491C (purple diamonds) decoding sites; wild-type bacterial ribosomes (black circles) are included for comparison. (B) Corresponding footprinting blots showing primer extensions starting with U1420. i, bacterial wild type; ii, mt wild type; iii, mt A1555G; iv, mt C1494T; v, bacterial G1491C.
When studying drug-induced miscoding, we found that the decoding accuracy of bacterial G1491C ribosomes is barely affected by aminoglycoside antibiotics. In absolute terms and in contrast to the mitochondrial A1555G and C1494T deafness mutants, the bacterial G1491C ribosomes showed little drug-induced misreading (see Fig. S2). The bacterial G1491C mutant and the hybrid deafness ribosomes differ primarily in 16S rRNA residues 1413 to 1415 and 1485 to 1487, which form the lower stem of helix 44 (see Fig. 1). At the structural level, helix 44 interacts with helix 27. By modeling, nucleotide alterations in the lower stem of H44 have been suggested to affect this interaction and the relative movement between these two helices as part of the conformational change required in decoding (30). Apparently, the nature of the lower stem plays an important role in both spontaneous and drug-aggravated miscoding and determines the translational accuracy of the mutant decoding sites. Thus, susceptibility of A1555G and C1494T deafness mitoribosomes is the result of two mechanisms, which act in concert: increased drug binding to its target and excessive aggravation of the mutants' inherent deficiency in ribosomal accuracy.
Several lines of evidence link aminoglycoside ototoxicity to the mitochondrial ribosome: (i) mitochondrial ribosomes are structurally more similar to their prokaryotic ancestor than to the eukaryotic cytosolic homologues; (ii) compared to cytosolic ribosomes, the mitochondrial ribosomes of higher eukaryotes exhibit a remarkable degree of aminoglycoside susceptibility (27) [see Table S2, which compares the drug susceptibility of hybrid bacterial ribosomes with the A site (H44) of human cytosolic ribosomes to that of hybrid ribosomes with the mitochondrial decoding site]; and (iii) idiosyncratic drug susceptibility is associated with genetic predisposition, in particular mutations in mtDNA: 20 to 40% of patients with aminoglycoside-induced ototoxicity either carry the A1555G or the C1494T mutation in the 12S rRNA gene (6, 40).
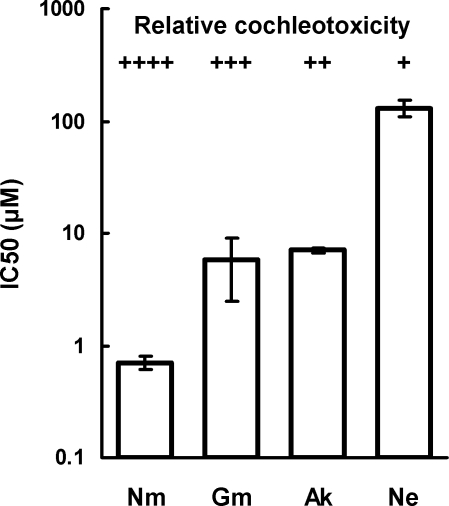
To further assess whether aminoglycoside-induced ototoxicity is a result of the drugs' anti-mitoribosomal activity, we compared the potencies of a series of aminoglycosides to inhibit mitoribosome function with their relative cochleotoxicity in humans (41). The correlation between these two measures (Fig. 4) is consistent with the hypothesis that aminoglycoside-induced cochleotoxicity relates to the drugs' activity against mitochondrial ribosomes. Further evidence for this hypothesis is provided by our finding that netilmicin, which displays the least cochlear toxicity of the clinical aminoglycosides (reviewed in ref. 42), is significantly less active against hybrid mitochondrial ribosomes than gentamicin, tobramycin, or amikacin.
Fig. 4.
Relationship between inhibition of protein synthesis in mitochondrial hybrid ribosomes and relative in vitro cochleotoxicity of aminoglycoside antibiotics. The potencies of a series of cochleotoxic aminoglycosides (Ak, amikacin; Gm, gentamicin; Ne, neamine; Nm, neomycin) in inhibiting protein synthesis in hybrid mitochondrial ribosomes (see Table 3) correlates with the relative cochleotoxicity previously reported by Kotecha and Richardson (41).
In summary, we provide experimental evidence for a mechanistic linkage between the mitochondrial A1555G and C1494T mutations and hypersusceptibility to aminoglycosides, although the exquisite tissue-specific action of aminoglycoside toxicity (that is, ototoxicity) is likely to involve additional factors (e.g., reactive oxygen species, drug uptake, or polyamine-like activation of NMDA receptors) (2, 43). Our results provide experimental support for aminoglycoside-induced dysfunction of the mitochondrial ribosome. We propose a pathogenic mechanism, in which interference of aminoglycosides with mitochondrial protein synthesis exacerbates the drugs' cochlear toxicity, playing a key role in sporadic dose-dependent and genetically inherited, aminoglycoside-induced deafness. Based upon our experiments, we suggest a scenario of aminoglycoside hearing loss, which is initiated by mitoribosomal misreading, subsequently via activation of downstream signaling pathways, such as MAPK and JNK (44, 45), misreading results in hair cell death through apoptosis.
Materials and Methods
Construction of Mutant Strains with Hybrid Ribosomes.
The recently described M. smegmatis mc2 155 SmS ΔrrnB (38) was used for all genetic manipulations. Site-directed mutagenesis of its single rRNA operon was done by PCR mutagenesis using hybrid rDNA oligonucleotides comprising the wild-type or mutant mitochondrial helix 44 decoding-site sequence. The resulting hybrid gene fragment was cloned into an integration-proficient plasmid used to transform M. smegmatis ΔrrnB. Transformants were selected on LB agar plates containing 20 μg/ml paromomycin for gene replacement by homologous recombination. Resulting recombinant M. smegmatis cells had the central 34-nucleotide part of the bacterial H44 replaced by its mitochondrial counterpart. Successful replacement of the bacterial decoding-site sequence with the mitochondrial sequence was controlled by sequence analysis of the chromosomal rrnA locus.
Minimal Inhibitory Concentration Assays.
Minimal inhibitory concentrations of neomycin B, paromomycin, kanamycin A, tobramycin, amikacin, gentamicin, netilmicin (all Sigma), and neamine were determined by broth microdilution assays as described previously (46). Neamine was a kind gift of Andrea Vasella, ETH Zurich. The gentamicin used in this study is a mixture of gentamicin C1, gentamicin C1a, and gentamicin C2 in a 45:35:30 ratio.
Isolation and Purification of Ribosomes.
Ribosomes were purified from bacterial cell pellets as described previously (30). In brief, ribosome particles were isolated by successive centrifugations and fractionated by sucrose gradient (10–40%) centrifugation. The 70S ribosome-enriched fraction was pelleted, resuspended in association buffer, incubated for 30 min at 4 °C, dispensed into aliquots, and stored at −80 °C following shock freezing in liquid nitrogen. Ribosome concentrations of 70S were determined by absorption measurements on the basis of 23 pmol ribosomes per A260 unit. Integrity and functional activity of purified 70S ribosomes was determined by analytical ultra-centrifugation and by assessing their capacity to form initiation complexes, as described previously (29).
Cell-free AUG(UUU)12 Translation Assays.
Cell-free translation reactions were done as described previously (30). A reaction mixture containing M. smegmatis tRNAbulk, amino acids, S100 extract, energy mix, pyruvate kinase, and polyamines was preincubated with 30 μM [14C]-phenylalanine (110 mCi/mmol) and/or 30 μM [3H]-leucine (500 mCi/mmol) at 37 °C for 15 min. The translation reaction was started by addition of ribosomes to a final concentration of 0.25 μM, AUG(UUU)12-mRNA (5′-GCGGCAAGGAGGUAAAUA AUG (UUU)12 UAA GCAGG-3′, obtained from Dharmacon) to 1 μM, and aminoglycoside antibiotics in serial dilutions. Following incubation for 60 min at 37 °C, the reaction was stopped by addition of KOH, precipitated polypeptides were collected on filters, and [14C]-phenylalanine or [3H]-leucine were quantified. Background values for Phe and Leu incorporation were 0.4 to 0.5 pmol at time zero; the background was not subtracted from the experimental values determined.
Cell-Free Luciferase Translation Assays.
Purified 70S hybrid ribosomes were used in a coupled transcription-translation reaction as described previously (29). The reaction mixture was incubated for 60 min at 37 °C, stopped on ice, and luciferase assay substrate (Promega) was added. Functional protein was quantified by measuring bioluminescence in a luminometer (Bio-Tek instruments, FLx800).
Footprinting Analyses.
DMS modification of 70S ribosomes (20 pmol) was performed in 100 μl buffer containing 80 mM potassium cacodylate (pH 6.5), 100 mM ammonium chloride, 20 mM magnesium chloride, 1 mM DTT, and 0.5 mM EDTA. Following ribosome activation for 15 min at 37 °C, gentamicin was added and the reaction mixture was incubated for another 15 min before addition of DMS (6 μl, 1:10 in ethanol). Following a 30-min incubation at 37 °C, the reaction was stopped by addition of 100 μl DMS Stop solution (50 mM Tris, pH 7.5, 300 mM sodium chloride, 1% SDS, 200 mM β-mercaptoethanol). Ribosomes were precipitated with ethanol, pelleted, resuspended in 200 μl 50 mM Tris pH 8, 0.5% SDS, and extracted with phenol/chloroform. DMS-modified RNA was precipitated with ethanol and sodium borohydride reduction and aniline-induced strand scission was performed as described previously (47). Primer extension of 16S rRNA was performed as described (48), using DNA oligonucleotides complementary to 16S-rRNA nucleotides 1445 to 1421. Air-dried gels were scanned and quantified using the STORM PhosphorImaging System with ImageQuant 5.2 Software (Amersham Bioscience).
Supplementary Material
Acknowledgments.
The authors thank Tanja Janušić (Institut für Medizinische Mikrobiologie) for expert technical assistance, Andrea Vasella (Swiss Federal Institute of Technology, Zurich) for kindly providing neamine, and Alexander Mankin for helpful comments on the manuscript. This work was supported by grants from the Swiss National Science Foundation (to E.C.B.) and from the Bonizzi-Theler-Stiftung (to S.N.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811258106/DCSupplemental.
References
- 1.Chambers HF, Sande MA. In: Goodman and Gilman's The Pharmacological Basis of Therapeutics. Goodman LS, Limbird LE, Milinoff PB, Gilman AG, Hardman JG, editors. New York: McGraw-Hill; 1996. pp. 1103–1121. [Google Scholar]
- 2.Forge A, Schacht J. Aminoglycoside antibiotics. Audiol Neurootol. 2000;5:3–22. doi: 10.1159/000013861. [DOI] [PubMed] [Google Scholar]
- 3.Matsui JI, Cotanche DA. Sensory hair cell death and regeneration: two halves of the same equation. Curr Opin Otolaryngol Head Neck Surg. 2004;12:418–425. doi: 10.1097/01.moo.0000136873.56878.56. [DOI] [PubMed] [Google Scholar]
- 4.Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 5.Fischel-Ghodsian N. Mitochondrial deafness mutations reviewed. Hum Mutat. 1999;13:261–270. doi: 10.1002/(SICI)1098-1004(1999)13:4<261::AID-HUMU1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 6.Zhao H, Li R, Wang Q, Yan Q, Deng JH. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Huang WG, Zha DJ, Qiu JH, Wang JL. Aspirin attenuates gentamicin ototoxicity: from the laboratory to the clinic. Hear Res. 2007;226:178–182. doi: 10.1016/j.heares.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Sha SH, Qiu JH, Schacht J. Aspirin to prevent gentamicin-induced hearing loss. N Engl J Med. 2006;354:1856–1857. doi: 10.1056/NEJMc053428. [DOI] [PubMed] [Google Scholar]
- 9.Hutchin T, Cortopassi G. Proposed molecular and cellular mechanism for aminoglycoside ototoxicity. Antimicrob Agents Chemother. 1994;38:2517–2520. doi: 10.1128/aac.38.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gale EF, Cundliffe E, Reynolds PE, Richmond MH, Waring JM. The Molecular Basis of Antibiotic Action. London: John Wiley & Sons, Inc.; 1981. [Google Scholar]
- 11.Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 12.Carter AP, Clemens WM, Brodersen DE, Morgan-Warren EJ, Wimberly BT. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 13.Böttger EC, Springer B, Prammananan T, Kidan Y, Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies J, Gorini L, Davis BD. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol Pharmacol. 1965;1:93–106. [PubMed] [Google Scholar]
- 15.Campuzano S, Vazquez D, Modolell J. Functional interaction of neomycin B and related antibiotics with 30S and 50S ribosomal subunits. Biochem Biophys Res Commun. 1979;87:960–966. doi: 10.1016/0006-291x(79)92050-3. [DOI] [PubMed] [Google Scholar]
- 16.Sander P, Prammananan T, Böttger EC. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol Microbiol. 1996;22:841–848. doi: 10.1046/j.1365-2958.1996.01532.x. [DOI] [PubMed] [Google Scholar]
- 17.Recht MI, Douthwaite S, Puglisi JD. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purohit P, Stern S. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature. 1994;370:659–662. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- 19.Griffey RH, Hofstadler SA, Sannes-Lowery KA, Ecker DJ, Crooke ST. Determinants of aminoglycoside-binding specificity for rRNA by using mass spectrometry. Proc Natl Acad Sci USA. 1999;96:10129–10133. doi: 10.1073/pnas.96.18.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shandrick S, et al. Monitoring molecular recognition of the ribosomal decoding site. Angew Chem Int Ed Engl. 2004;43:3177–3182. doi: 10.1002/anie.200454217. [DOI] [PubMed] [Google Scholar]
- 21.Kaul M, Barbieri CM, Pilch DS. Aminoglycoside-induced reduction in nucleotide mobility at the ribosomal RNA A-site as a potentially key determinant of antibacterial activity. J Am Chem Soc. 2006;128:1261–1271. doi: 10.1021/ja056159z. [DOI] [PubMed] [Google Scholar]
- 22.Kondo J, Westhof E. The bacterial and mitochondrial ribosomal A-site molecular switches possess different conformational substates. Nucleic Acids Res. 2008;36:2654–2666. doi: 10.1093/nar/gkn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu DH, Rando RR. Aminoglycoside binding to human and bacterial A-Site rRNA decoding region constructs. Bioorg Med Chem. 2001;9:2601–2608. doi: 10.1016/s0968-0896(01)00034-7. [DOI] [PubMed] [Google Scholar]
- 24.Hamasaki K, Rando RR. Specific binding of aminoglycosides to a human rRNA construct based on a DNA polymorphism which causes aminoglycoside-induced deafness. Biochemistry. 1997;36:12323–12328. doi: 10.1021/bi970962r. [DOI] [PubMed] [Google Scholar]
- 25.Kaul M, Barbieri CM, Pilch DS. Defining the basis for the specificity of aminoglycoside-rRNA recognition: a comparative study of drug binding to the A sites of Escherichia coli and human rRNA. J Mol Biol. 2005;346:119–134. doi: 10.1016/j.jmb.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 26.Pfister P, et al. Mutagenesis of 16S rRNA C1409–G1491 base-pair differentiates between 6′OH and 6′NH3+ aminoglycosides. J Mol Biol. 2005;346:467–475. doi: 10.1016/j.jmb.2004.11.073. [DOI] [PubMed] [Google Scholar]
- 27.Kurtz DI. Fidelity of protein synthesis with chicken embryo mitochondrial and cytoplasmic ribosomes. Biochemistry. 1974;13:572–577. doi: 10.1021/bi00700a026. [DOI] [PubMed] [Google Scholar]
- 28.Ryu DH, Rando RR. Decoding region bubble size and aminoglycoside antibiotic binding. Bioorg Med Chem Lett. 2002;12:2241–2244. doi: 10.1016/s0960-894x(02)00342-6. [DOI] [PubMed] [Google Scholar]
- 29.Hobbie SN, et al. Engineering the rRNA decoding site of eukaryotic cytosolic ribosomes in bacteria. Nucleic Acids Res. 2007;35:6086–6093. doi: 10.1093/nar/gkm658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hobbie SN, et al. Mitochondrial deafness alleles confer misreading of the genetic code. Proc Natl Acad Sci USA. 2008;105:3244–3249. doi: 10.1073/pnas.0707265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Stasio EA, Moazed D, Noller HF, Dahlberg AE. Mutations in 16S ribosomal RNA disrupt antibiotic-RNA interactions. EMBO J. 1989;8:1213–1216. doi: 10.1002/j.1460-2075.1989.tb03494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Stasio EA, Dahlberg AE. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16 S ribosomal RNA. J Mol Biol. 1990;212:127–133. doi: 10.1016/0022-2836(90)90309-A. [DOI] [PubMed] [Google Scholar]
- 33.Prammananan T, Sander P, Springer B, Böttger EC. RecA-mediated gene conversion and aminoglycoside resistance in strains heterozygous for rRNA. Antimicrob Agents Chemother. 1999;43:447–453. doi: 10.1128/aac.43.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Recht MI, Puglisi JD. Aminoglycoside resistance with homogeneous and heterogeneous populations of antibiotic-resistant ribosomes. Antimicrob Agents Chemother. 2001;45:2414–2419. doi: 10.1128/AAC.45.9.2414-2419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfister P, Hobbie S, Vicens Q, Böttger EC, Westhof E. The molecular basis for A-Site mutations conferring aminoglycoside resistance: relationship between ribosomal susceptibility and X-ray crystal structures. ChemBioChem. 2003;4:1078–1088. doi: 10.1002/cbic.200300657. [DOI] [PubMed] [Google Scholar]
- 36.Hobbie SN, Pfister P, Brull C, Westhof E, Böttger EC. Analysis of the contribution of individual substituents in 4,6-aminoglycoside-ribosome interaction. Antimicrob Agents Chemother. 2005;49:5112–5118. doi: 10.1128/AAC.49.12.5112-5118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hobbie SN, et al. Binding of neomycin-class aminoglycoside antibiotics to mutant ribosomes with alterations in the A-site of 16S rRNA. Antimicrob Agents Chemother. 2006;50:1489–1496. doi: 10.1128/AAC.50.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hobbie SN, et al. A genetic model to investigate structural drug-target interactions at the ribosomal decoding site. Biochimie. 2006;88:1033–1043. doi: 10.1016/j.biochi.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 39.Vicens Q, Westhof E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure (Camb) 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- 40.Estivill X, et al. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment of aminoglycosides. Am J Hum Genet. 1998;62:27–35. doi: 10.1086/301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotecha B, Richardson GP. Ototoxicity in vitro: effects of neomycin, gentamicin, dihydrostreptomycin, amikacin, spectinomycin, neamine, spermine and poly-L-lysine. Hear Res. 1994;73:173–184. doi: 10.1016/0378-5955(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 42.Begg EJ, Barclay ML. Aminoglycosides-50 years on. Br J Clin Pharmacol. 1995;39:597–603. [PMC free article] [PubMed] [Google Scholar]
- 43.Basile AS, et al. N-methyl-D-aspartate antagonists limit aminoglycoside antibiotic-induced hearing loss. Nat Med. 1996;2:1338–1343. doi: 10.1038/nm1296-1338. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, et al. A peptide inhibitor of c-Jun N-terminal kinase protects against both aminoglycoside and acoustic trauma-induced auditory hair cell death and hearing loss. J Neurosci. 2003;23:8596–8607. doi: 10.1523/JNEUROSCI.23-24-08596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalinec GM, et al. Pivotal role of Harakiri in the induction and prevention of gentamicin-induced hearing loss. Proc Natl Acad Sci USA. 2005;102:16019–16024. doi: 10.1073/pnas.0508053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfister P, et al. 23S rRNA base pair 2057–2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A->G. Proc Natl Acad Sci USA. 2005;102:5180–5185. doi: 10.1073/pnas.0501598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peattie DA. Direct chemical method for sequencing RNA. Proc Natl Acad Sci USA. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merryman C, Noller HF. In: RNA:Protein Interactions: A Practical Approach. Smith CWJ, editor. Oxford, UK: Oxford Univ Press; 1998. pp. 237–253. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.