Abstract
SET domain protein lysine methyltransferases (PKMTs) regulate transcription and other cellular functions through site-specific methylation of histones and other substrates. PKMTs catalyze the formation of monomethylated, dimethylated, or trimethylated products, establishing an additional hierarchy with respect to methyllysine recognition in signaling. Biochemical studies of PKMTs have identified a conserved position within their active sites, the Phe/Tyr switch, that governs their respective product specificities. To elucidate the mechanism underlying this switch, we have characterized a Phe/Tyr switch mutant of the histone H4 Lys-20 (H4K20) methyltransferase SET8, which alters its specificity from a monomethyltransferase to a dimethyltransferase. The crystal structures of the SET8 Y334F mutant bound to histone H4 peptides bearing unmodified, monomethyl, and dimethyl Lys-20 reveal that the phenylalanine substitution attenuates hydrogen bonding to a structurally conserved water molecule adjacent to the Phe/Tyr switch, facilitating its dissociation. The additional space generated by the solvent's dissociation enables the monomethyllysyl side chain to adopt a conformation that is catalytically competent for dimethylation and furnishes sufficient volume to accommodate the dimethyl ε-ammonium product. Collectively, these results indicate that the Phe/Tyr switch regulates product specificity through altering the affinity of an active-site water molecule whose dissociation is required for lysine multiple methylation.
Keywords: enzyme mechanism, histone methylation, transcription, chromatin, enzyme kinetics
Lysine methylation is a widespread covalent modification that occurs in histones and nonhistone proteins. Site-specific methylation of lysines in histones H3, H4, and H1b has been implicated in a myriad of functions, including transcriptional regulation, heterochromatin formation, and DNA damage response (1). Various effector proteins mediate these functions through sequence-specific recognition of lysine methyl marks. In addition, the lysine ε-amine group can be monomethylated, dimethylated, or trimethylated, imparting a further level of specificity in methyllysine signaling. The importance of this specificity has been emphasized by recent studies illustrating that specific methylation states are enriched in distinct regions of genes (2). Correlatively, certain methyllysine binding domains discriminate among different degrees of lysine methylation (3), underscoring the synergy between the site and degree of methylation in signaling.
A key to deciphering the functions of lysine methylation is to understand the specificities of the enzymes that establish these marks. Structural studies of PKMTs belonging to the SET domain family have yielded insights into the mechanisms by which these enzymes recognize and methylate specific sites within their cognate substrates (4). Protein substrates generally bind in a β-strand conformation, depositing the lysyl side chain in a channel that traverses the core of the catalytic domain and links to the S-adenosyl-l-methionine (AdoMet) binding site. Mutational analyses have revealed a specific site within the lysine binding channel that modulates the product specificities of histone methyltransferases (Fig. S1). In lysine monomethyltransferases, this position is occupied by a tyrosine, whereas in most dimethyltransferases and trimethyltransferases, a phenylalanine or another hydrophobic amino acid is located in this site (5–10). These findings underpin a Phe/Tyr switch model for product specificity wherein the identity of the residue occupying the switch position differentiates monomethyltransferases and di/trimethyltransferases, with the exception of enzymes that are active within the context of heteromeric complexes whose subunits regulate product specificity, such as ScSET1 (11) and HsMLL (12, 13). Despite its biological importance, the molecular mechanism underlying the Phe/Tyr switch is poorly understood. Indeed, this reflects a fundamental question: how are methyl groups of monomethyllysine, dimethyllysine, and trimethyllysine physically arranged in the active sites of PKMTs during successive rounds of methylation? This question has remained unaddressed because of a lack of structures of SET domain PKMTs bound to lysine substrates in different methylation states.
To resolve these issues and gain mechanistic insights into SET domain product specificity, we have structurally and functionally characterized a Phe/Tyr switch mutant of the H4K20-specific monomethyltransferase SET8 (also known as PR-SET7 and KMT5A). We previously reported that a phenylalanine mutation of the Phe/Tyr switch residue Tyr-334 alters the product specificity of SET8 to a dimethyltransferase, but does not affect histone H4 binding or the overall methylation rate (6). We chose this mutant for further analysis to evaluate how a subtle mutation from tyrosine to phenylalanine can fundamentally alter product specificity. Crystal structures of SET8 Y334F in complex with a histone H4 peptide bearing an unmodified (H4K20), monomethylated (H4K20me1), and dimethylated Lys-20 (H4K20me2) reveal that the Phe/Tyr switch modulates the affinity of a structurally conserved water molecule within the active site. Dissociation of this water molecule permits a monomethyllysine substrate to adopt an alternative conformation conducive for dimethylation. Together, these findings demonstrate that the Phe/Tyr switch governs SET domain product specificity through the intermediacy of a water molecule.
Results
Kinetic Characterization of SET8 Y334F Mutant.
In our initial studies, we reported the kinetic parameters of the SET8 Y334F mutant (residues 191–352) by using a 10-residue H4K20 peptide substrate (KM = 520 μM, kcat = 0.45 min−1), representing the aggregate rates of monomethylation and dimethylation (6). To measure the individual rate constants for these reactions, SET8 Y334F was assayed with an H4K20me1 peptide and a H4K20 peptide by using short assay times that exclude the formation of the dimethylated product. Under these conditions, SET8 Y334F displays comparable KM values for the 2 peptides and similar binding affinities, as measured by isothermal titration calorimetry, but methylates the H4K20 peptide more efficiently than H4K20me1 (Table 1). The kcat value for the H4K20me1 peptide is consistent with the rate constants for dimethylation and trimethylation of an H3K27 peptide by a viral SET domain methyltransferase (vSET) (7) and for the methylation of an H3K9me2 peptide by SUV39H1 (14). Because of its comparable affinities for H4K20 and H4K20me1, these results imply that the chemistry of the dimethylation reaction may be rate-limiting in SET8 Y334F (see the final paragraph in Discussion).
Table 1.
Biochemical analysis of native SET8 and the Y334F mutant
| SET8 | Peptide substrate | KD, μM | KM, μM | kcat, min−1 | kcat/KM, μM−1 × min−1 × 103 |
|---|---|---|---|---|---|
| Native | H4K20* | 33 ± 1 | 400 ± 30 | 0.43 ± 0.02 | 1.1 ± 0.1 |
| H4K20me1 | 24 ± 1 | NA | NA | NA | |
| Y334F† | H4K20 | 19 ± 2* | 490 ± 40 | 0.43 ± 0.09 | 0.88 ± 0.20 |
| H4K20me1 | 32 ± 1 | 260 ± 30 | 0.016 ± 0.001 | 0.062 ± 0.008 |
NA, no activity detected.
*Data previously reported and shown for comparative purposes (6). [Reproduced with permission from ref. 6 (Copyright 2005, Cold Spring Harbor Lab Press).]
†No heat of binding was detected between SET8 Y334F and the H4K20me2 peptide by isothermal titration calorimetry (ITC). Error values were calculated from the nonlinear fits to the kinetic and ITC data.
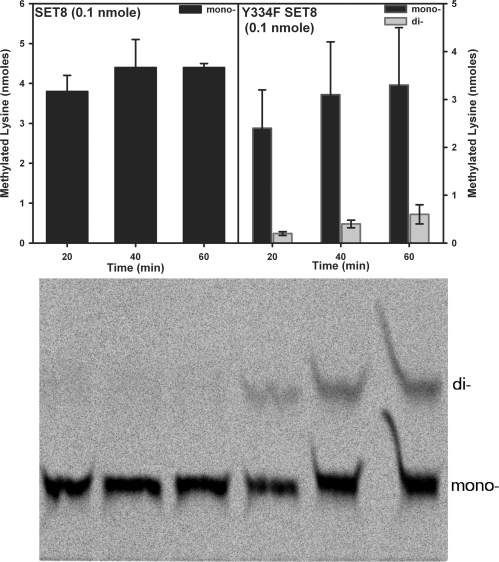
To define whether SET8 Y334F uses a processive or distributive kinetic mechanism, the amounts of monomethyllysine and dimethyllysine products were quantitatively measured by using TLC (Fig. 1). In processive methylation, monomethyllysine is a reaction intermediate that remains bound to the enzyme and thus cannot exceed the total enzyme concentration, whereas in a distributive mechanism, the monomethyllysine is released into solution where it accumulates at concentrations that exceed the enzyme before its complete conversion to the dimethylated product. Incubation of a limited amount of SET8 Y334F (0.1 nmol) with the H4K20 peptide and AdoMet yielded 4.4 nmol of H4K20me1 and 0.8 nmol of H4K20me2 in 1 h, consistent with a distributive mechanism for this mutant.
Fig. 1.
Analysis of the product specificities of native SET8 and the Y334F mutant. TLC-based assays of SET8 and the Y334F mutant with the unmodified H4K20 peptide and 3H-AdoMet as substrates are illustrated. The monomethyllysine and dimethyllysine products generated by native SET8 and the Y334F mutant were separated on a TLC plate (Lower), extracted, quantified, and displayed as a bar graph (Upper).
Structures of SET8 Y334F Bound to H4K20, H4K20me1, and H4K20me2.
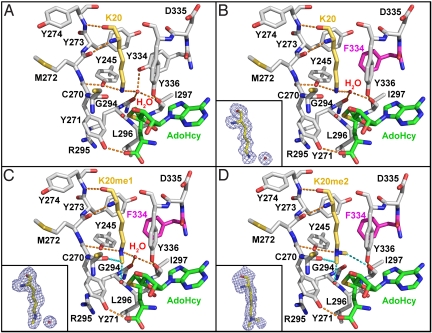
To elucidate the mechanism by which the Y334F mutation alters the product specificity of SET8, we solved the crystal structures of this mutant in ternary complexes with H4K20, H4K20me1, and H4K20me2 peptides and the product S-adenosyl-l-homocysteine (AdoHcy) (Table S1). The high resolution of the diffraction data (1.25- to 1.6-Å resolution) permitted unambiguous modeling of the unmodified and methylated K20 side chains in the omit maps (Fig. 2). The structures of the SET8 Y334F complexes are highly similar to the native enzyme and display the same network of interactions between the H4 peptide and the enzyme (6, 8). These observations indicate that the Y334F mutation does not induce major conformational changes in the enzyme, implying that the altered product specificity is caused by local rearrangements in the active site.
Fig. 2.
Structures of the active sites of native SET8 and the Y334F mutant bound to AdoHcy and H4K20, H4K20me1, and H4K20me2 peptides. His-18 in histone H4 forms 1 wall of the channel and was omitted to provide an unobstructed view of the active site. SET8, histone H4, and AdoHcy are delineated by gray, gold, and green carbon atoms, respectively. The Y334F mutation is highlighted in magenta. (Insets) Shown are the FO − FC omit map electron densities for the various K20 side chains contoured at 2.0 σ. Conventional and CH…O hydrogen bonds are depicted as orange and cyan dashes, respectively. (A) Native SET8–H4K20–AdoHcy complex (Protein Data Bank ID code 1ZKK). (B) SET8 Y334F–H4K20–AdoHcy. (C) SET8 Y334F–H4K20me1–AdoHcy. (D) SET8 Y334F–H4K20me2–AdoHcy.
A comparison of the active-site structures of native SET8 and the Y334F mutant bound to the H4K20 peptide (Fig. 2 A and B) illustrates that the K20 side chain is bound in an all-trans extended conformation within a channel composed of 4 aromatic residues, Tyr-245, the Phe/Tyr switch residue 334, and Tyr-336 in SET8, and His-18 in histone H4 (residues not shown). This channel connects to the AdoMet binding pocket via an oxygen-rimmed methyltransfer pore through which a methyl group transits during catalysis (15). In both native SET8 and the Y334F mutant, the conformation of the K20 side chain is oriented via hydrogen bonding to the Tyr-245 OH group and a tightly bound water molecule that occupies a cleft in the active site that we refer to as the solvent pocket. In the native enzyme, the water molecule is further coordinated within this pocket via hydrogen bonds to the Tyr-334 OH group and the carbonyl oxygens of Gly-294 and Ile-297 (Fig. 2A), whereas in SET8 Y334F, the Tyr-334 hydrogen bond to the water is absent because of the phenylalanine mutation (Fig. 2B). The structures of other SET domain PKMTs, including SET7/9, SUV39H2, G9A, and G9A-like protein (GLP), also possess solvent binding pockets and a water molecule (Fig. S2 A–E), suggesting their involvement in substrate binding or catalysis.
Similar to the H4K20-bound complexes, the K20me1 side chain adopts an extended all-trans configuration in the lysine binding channel of the Y334F mutant (Fig. 2C). The backbone atoms of H4K20me1 and H4V21 are shifted 0.5 Å away from the center of the active site compared with the SET8 Y334F-H4K20 complex to accommodate the K20me1 side chain. However, this displacement does not alter the overall histone H4 binding mode, nor disrupt hydrogen bonding to the H4K20me1 ε-amine. The interactions orient the K20me1 side chain in a posttransfer product conformation with its methyl group linearly aligned with the AdoHcy thioether moiety, mimicking the monomethyllysyl binding modes observed in native SET8 and SET7/9 product complexes (Fig. S2 E and F) (8, 9, 16–18).
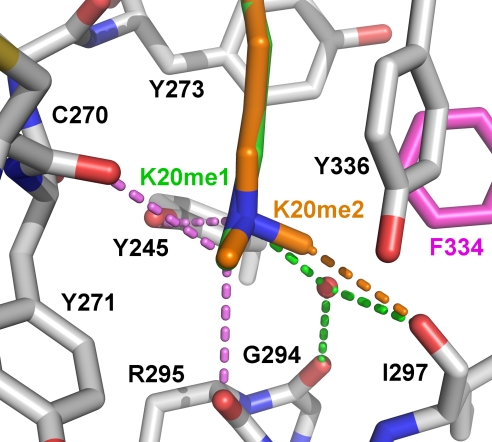
In contrast, the structure of the SET8 Y334F bound to the H4K20me2 peptide reveals variable dimethyllysyl binding modes. In 1 molecule in the unit cell, the K20me2 side chain adopts an extended all-trans conformation, whereas in the other complexes, the side chain assumes either non-trans or kinked configurations. In the kinked orientation, a rotation along the Cδ–Cε bond in the K20me2 side chain shifts the dimethyl ε-amine toward residues Cys-270–Met-272 in the lysine binding channel, generating a void adjacent to the methyltransfer pore that is occupied by water molecules (Fig. S3). In contrast, the all-trans K20me2 side chain extends the length of the lysine binding channel, excluding water molecules from the active site (Fig. 2D). We conclude that this conformation represents a bona fide dimethyllysine product complex because: (i) the H4K20me1 and H4K20me2 side chains are superimposable (Fig. 3), (ii) hydrogen bonding is maintained between the H4K20me2 ε-amine and Tyr-245, and (iii) a superimposition of this complex and the GLP–H3K9me2–AdoHcy product complex illustrates identical dimethyllysyl side-chain conformations (Fig. S4). An important consequence of this all-trans binding mode is the displacement of the active-site water molecule from the solvent pocket by the dimethyl ε-amine (Fig. 2D). Specifically, 1 of the K20me2 methyl groups projects into the vacant solvent binding site, forming a 3.3-Å carbon-oxygen (CH···O) hydrogen bond with the carbonyl oxygen of Ile-297 (Fig. 3). Similarly, the K9me2 side chain adopts an identical conformation and engages in the same interactions in the GLP–H3K9me2 product complex (Fig. S4). The presence of CH···O hydrogens bonds in methyllysine recognition concurs with prior studies of SET domain PKMTs (15, 19) and explains the capacity of the SET8 Y334F active site to accommodate a dimethyllysine. Specifically, the affinity for the water molecule bound within the solvent pocket is diminished through the loss of a hydrogen bond because of the Y334F mutation, enabling a methyl group to compete for binding within the pocket via a compensatory CH···O hydrogen bond.
Fig. 3.
Monomethyllysine and dimethyllysine binding by SET8 Y334F. H4K20me1 (green carbons) and H4K20me2 (orange carbons) are superimposed within the active site. Hydrogen bonds between the active site and the K20me1 and K20me2 side chains are illustrated according to the corresponding colors of the carbon atoms in their side chains, and hydrogen bonds common to both complexes are shown in violet.
Mechanism of Lysine Monomethylation and Dimethylation.
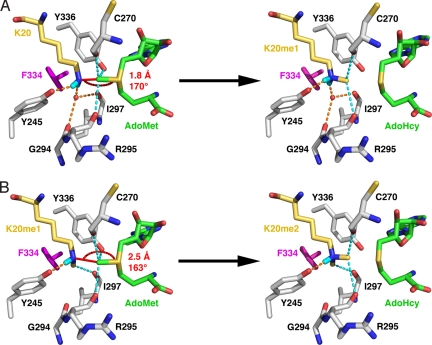
Based on the structures of the SET8 Y334F complexes, we propose a mechanism for lysine dimethylation applicable to other SET domain PKMTs. Modeling of a SET8 Y334F–AdoMet–H4K20 substrate complex illustrates that hydrogen bonds to the Tyr-245 OH group and the active-site water molecule align the deprotonated K20 ε-amine and the AdoMet methyl group and sulfonium cation with a reaction distance of 1.8 Å and an angle of 170°, consistent with a linear SN2 methyltransfer reaction (Fig. 4A). The Tyr-245 OH group, which is conserved in most histone methyltransferases (Fig. S1), is critical for the productive alignment of the K20 ε-amine group, because mutations of this residue abolish methylation (6). This result is supported by structural analysis of the SET8 Y245F–H4K20–AdoHcy complex in which the K20 side chain adopts a kinked conformation with its ε-amine misaligned for methyltransfer (Fig. S5).
Fig. 4.
Model for H4K20 dimethylation by the SET8 Y334F mutant. (A) First methyltransfer reaction. In the SET8 Y334F–H4K20–AdoMet complex, AdoMet was modeled in the active site by using the AdoHcy coordinates (Fig. 2 color scheme with hydrogens rendered in cyan). The SN2 reaction distance and bond angle corresponding to the H4K20 ε-amine group and the AdoMet methyl group and sulfonium cation are noted. Fig. S6 illustrates the hydrogen bonding to the K20 ε-amine. (B) Second methyltransfer reaction. In this substrate complex, the H4K20me1 side chain is modeled with its methyl group projecting into the vacated water binding site, which is inferred from the coordinates of the corresponding methyl group in the H4K20me2 product complex (Fig. 2D). This orientation aligns the K20me1 ε-amine for methyltransfer with AdoMet.
After the first methyltransfer reaction, the K20me1 side chain is protonated and adopts an all-trans geometry with its methyl group oriented toward the methyltransfer pore (Fig. 4A and Fig. S6A). In monomethyltransferases, this configuration represents a terminal product complex stabilized by the water molecule coordinated by 4 hydrogen bonds within the solvent pocket (Fig. S2 E and F). For dimethylation to occur, the monomethyllysyl side chain must adopt a new conformation wherein its methyl group is rotated out of the transfer path, and its ε-amine is deprotonated to function as a nucleophile. The SET8 Y334F–H4K20me2 complex (Fig. 2D) yields insights as to how these criteria are met. In dimethyltransferases and trimethyltransferases, only 3 hydrogen bonds coordinate the water molecule within the solvent binding pocket, diminishing its affinity for the active site. Upon the water's dissociation, the monomethyllysyl side chain can undergo a conformational change by rotation along its Cε–Nζ bond, repositioning the methyl group in the vacant solvent pocket where it forms a CH···O hydrogen bonds with Ile-297 that stabilizes this alternative conformation (Fig. 4B and Fig. S6B). This realignment maintains hydrogen bonding between the K20me1 ε-amine and Tyr-245 while rotating the methyl group out of the transfer path. The rotation of the K20me1 side chain also breaks 1 of the 2 hydrogen bonds to the ε-ammonium ion, facilitating its deprotonation before the second methyltransfer reaction. Modeling of the SET8 Y334–H4K20me1–AdoMet substrate complex illustrates that the deprotonated H4K20me1 ε-amine and AdoMet methyl group align with a geometry of 2.5 Å and 163°. This distance is longer than the value calculated for the first reaction, caused in part by the aforementioned 0.5-Å shift of K20me1 and K20me2 away from the center of the active site. Taking this shift into account, the distance between the methyl group and ε-amine is ≈2.0 Å, comparable to the reaction distance for monomethylation (Fig. 4A). In the final product complex, the dimethyllysyl side chain adopts an all-trans geometry, as observed in the SET8 Y334F–H4K20me2 and GLP–H3K9me2 complexes (Fig. S4)
Discussion
The mechanisms underlying the product specificity of SET domain PKMTs have remained the subject of intense study because of the important regulatory roles of lysine methylation in transcription and other cellular functions. Initial structural studies attributed differences in product specificity to several factors, including steric hindrance by the tyrosine hydroxyl group in monomethyltransferases (10) and variations in the diameter of the lysine binding channel caused by the size of residue occupying Phe/Tyr switch site (7–9, 19). We evaluated these models in light of our results indicating that the Phe/Tyr switch regulates product specificity through the active-site water molecule. Docking of the H4K20me2 peptide into the substrate binding cleft of native SET8 reveals that the K20me2 methyl group that binds in the solvent pocket is positioned 1.5 Å from the active-site water and 2.7 Å from the Tyr-334 OH group (Fig. S7). The distance between the water and the methyl group is indicative of a severe clash that would inhibit H4K20 dimethylation, in agreement with the product specificity of native SET8. Although the methyl group and Tyr-334 OH group are in relatively close proximity, this distance does not necessarily represent steric hindrance because CH···O hydrogen bonds of comparable lengths (2.7–3.2 Å) have been observed with methyllysines in the active sites of SET domain PKMTs and a JmjC lysine demethylase (15, 20). Additionally, we investigated how changes in the lysine binding channel's diameter may influence product specificity. The Y334F substitution results in a modest widening at the channel's midpoint based on a comparison of native SET8 and the Y334F mutant (Fig. S8 A and B). However, the diameter at the base of the channel adjacent to the methyltransfer pore remains narrow owing to the bound water molecule. Dissociation of the water expands the channel's diameter by unmasking the solvent pocket, yielding sufficient volume to accommodate dimethylation by SET8 Y334F (Fig. S8C). Collectively, these findings demonstrate that diffusion of the water molecule from the active site relieves steric hindrance and enlarges the diameter of the lysine binding channel's base, facilitating dimethylation.
Recently, a series of quantum mechanical and molecular dynamic studies have posited different theoretical models for SET domain product specificity. Initial simulations of lysine methylation by SET7/9 ascribed its product specificity to variations in near attack conformations (21), whereas more recent modeling studies by Zhang and coworkers (22) favor a model wherein chains of water molecules hydrogen bond to the deprotonated methyllysine ε-amine, inhibiting dimethylation and trimethylation. Conversely, a series of simulations with multiple SET domain PKMTs show that solvent chains are responsible for deprotonating the lysine ε-amine to promote methyltransfer (23–27), whereas the results of another modeling study of SET7/9 implicate the invariant active site tyrosine (Tyr-335; Figs. S1 and S2) as the catalytic base in deprotonation (28). Although there is no experimental data for solvent chains in our SET8 structures or in other SET domain PKMTs, it is conceivable that these chains could form transiently in solution to provide a deprotonation path during catalysis, in agreement with our model (see below). With respect to the invariant tyrosine, structural and computational analyses have shown that the distance and angle between its hydroxyl group and the lysine ε-ammonium cation are unfavorable for proton abstraction (7, 19) and that the theoretical pKa value of its hydroxyl group (>13.0) is inconsistent with base catalysis (23, 27).
We propose that our model for the product specificity of SET8 is applicable to the SET domain family because of the structural homology of the lysine binding channel, the solvent pocket, and the methyllysine conformations observed in the structures of other PKMTs (Fig. S2). The conserved binding mode of the active-site water and its hydrogen bonding to the lysine substrate imply that it functions as a molecular placeholder during catalysis. In monomethylation, the water molecule hydrogen bonds to the lysine ε-amine, aligning it in a linear geometry with the AdoMet methyl group (Fig. 4A). However, the hydrogen bonding between the water and the monomethyllysine restricts further rounds of methylation by impeding the rotation and deprotonation of the monomethyl ε-ammonium group. Dissociation of the water in dimethyltransferases and trimethyltransferases relieves these inhibitory effects, enabling the methyl group to rotate into the vacant solvent pocket to orient the deprotonated ε-amine for the second methyltransfer (Fig. 4B).
In addition to its placeholder role, the active-site water may facilitate deprotonation of the monomethyllysine ε-ammonium cation during dimethylation. Theoretical calculations predict that the pKa values of the lysine or methyllysine ε-ammonium cations are depressed to 8.0–8.2 because of the hydrophobicity of the lysine binding channel (23, 25, 27), consistent with the pKa values for kcat derived from pH rate profiles of SET domain PKMTs (29, 30). These ionization constants concur with water-mediated deprotonation of the ε-ammonium cation, which could occur via solvent entering the active site during cofactor exchange between turnovers or via water molecule chains that form within the active site of the Michaelis complex (23–27). Alternatively, the active-site water molecule serves as a hydrogen-bond acceptor with H4K20me1 (Fig. 4A) and could directly deprotonate the ε-ammonium cation before dimethylation, dissociating from the active site as a hydronium ion.
These findings also provide a rationale for the decrease in rate constants associated with dimethylation and trimethylation reported for various SET domain PKMTs. SET8 Y334F methylates the H4K20me1 peptide >20-fold more slowly than that for the H4K20 peptide while exhibiting comparable affinity for both substrates (Table 1). These results imply that a chemical step or a conformational change in the dimethylation reaction is rate-limiting and, in conjunction with the structural data, suggest that the dissociation of the active-site water molecule and concomitant realignment and deprotonation of the K20me1 side chain represent the rate-limiting steps in lysine dimethylation by SET8 Y334F. Correlatively, the reported rate constants for dimethylation and trimethylation of other SET domain PKMTs, such as vSET, G9A, SUV39H1, and Rubisco large-subunit methyltransferase are reduced 3- to 10-fold compared with their respective rates of monomethylation (7, 14, 29, 31), implying that methyllysine reorientation and deprotonation between turnovers constitute a common rate-limiting step in catalysis. This kinetic model is applicable to PKMTs that catalyze processive methylation in which the protein substrate remains bound during multiple turnovers and to enzymes that obey a distributive mechanism wherein the methylated substrate dissociates and rebinds to the enzyme with the methyllysine in a productive alignment for subsequent methylation. Future studies are needed to illuminate the conformational changes that facilitate lysine trimethylation by PKMTs.
Experimental Procedures
Synthetic Peptides.
H4K20, H4K20me1, and H4K20me2 peptides (sequence: A15KRHRK20VLRD24) were purchased from New England Peptide and were suspended in either calorimetry or gel filtration buffer before use (6). The H4K20 peptide used in the TLC assays was synthesized with a C-terminal biotinylated lysine for avidin-based purification (sequence: A15KRHRK20VLRDN25K-biotin).
Protein Expression and Crystallization.
The Y334F and Y245F mutants of human SET8 (residues 191–352) were overexpressed and purified as reported (6). Concentrated enzyme (40 mg/mL) was mixed with a 2-fold-molar excess of AdoHcy and the H4 peptides for crystallization. Crystals were obtained in 23–40% pentaerythritol ethoxylate (15/4), 50 mM (NH4)2SO4, and 50 mM Bis-Tris, pH 6.5–7.0 at 4 °C and were harvested as published (6).
Data Collection and Structure Determination.
Diffraction data were collected at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL) and the European Synchrotron Radiation Facility (Grenoble, France) and were processed with HKL2000 (32) or XDS (33). Structures of the SET8 mutants were solved by molecular replacement using MOLREP (34) and the coordinates of the SET8–H4K20–AdoHcy complex (Protein Data Bank ID code 1ZKK). The structures were refined with REFMAC (35), and the models were manually adjusted by using O (36). Crystallographic data and refinement statistics are reported in Table S1. Structural figures were rendered with PyMOL (http://pymol.sourceforge.net).
Calorimetry and Methyltransferase Assays.
Equilibrium dissociation (KD) constants (Table 1) were measured with a VP-ITC calorimeter (MicroCal) as reported (6). The binding isotherms were plotted by using ORIGIN software package (OriginLab) and displayed binding stoichiometries between 0.9 and 1.0. Kinetic characterization of the SET8 Y334F was performed in duplicate by using a fluorescent assay as described (6, 37) with the exception that 1.0–3.0 μM SET8 Y334F and 200 μM AdoMet were used in the assays. TLC was performed as outlined in SI Text.
Supplementary Material
Acknowledgments.
We thank Laurence Serre at the FIP beamline BM30A at the European Synchrotron Research Facility for assistance with data collection. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-ENG-38. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago. Data were also collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source. This work was supported by National Institutes of Health Grant GM073839 (to R.C.T.), Department of Energy Grant DE-FG02-92ER20075 (to R.L.H.), and a Canadian Institutes of Health Research Postdoctoral Fellowship (to J.-F.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3F9W, 3F9X, 3F9Y, and 3F9Z).
This article contains supporting information online at www.pnas.org/cgi/content/full/0806712105/DCSupplemental.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: Protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins RE, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couture JF, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev. 2005;19:1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian C, et al. Structural insights of the specificity and catalysis of a viral histone H3 lysine 27 methyltransferase. J Mol Biol. 2006;359:86–96. doi: 10.1016/j.jmb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Xiao B, et al. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 2005;19:1444–1454. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao B, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider J, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 13.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 14.Chin HG, Patnaik D, Estève PO, Jacobsen SE, Pradhan S. Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: Participation of the chromodomain in enzymatic catalysis. Biochemistry. 2006;45:3272–3284. doi: 10.1021/bi051997r. [DOI] [PubMed] [Google Scholar]
- 15.Couture JF, Hauk G, Thompson MJ, Blackburn GM, Trievel RC. Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases. J Biol Chem. 2006;281:19280–19287. doi: 10.1074/jbc.M602257200. [DOI] [PubMed] [Google Scholar]
- 16.Chuikov S, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 17.Couture JF, Collazo E, Hauk G, Trievel RC. Structural basis for the methylation site specificity of SET7/9. Nat Struct Mol Biol. 2006;13:140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian K, et al. Regulation of estrogen receptor α by the SET7 lysine methyltransferase. Mol Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trievel RC, Flynn EM, Houtz RL, Hurley JH. Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat Struct Biol. 2003;10:545–552. doi: 10.1038/nsb946. [DOI] [PubMed] [Google Scholar]
- 20.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 21.Hu P, Zhang Y. Catalytic mechanism and product specificity of the histone lysine methyltransferase SET7/9: An ab initio QM/MM-FE study with multiple initial structures. J Am Chem Soc. 2006;128:1272–1278. doi: 10.1021/ja056153+. [DOI] [PubMed] [Google Scholar]
- 22.Hu P, Wang S, Zhang Y. How do SET-domain protein lysine methyltransferases achieve the methylation state specificity? Revisited by Ab initio QM/MM molecular dynamics simulations. J Am Chem Soc. 2008;130:3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Bruice TC. Histone lysine methyltransferase SET7/9: Formation of a water channel precedes each methyl transfer. Biochemistry. 2007;46:14838–14844. doi: 10.1021/bi7014579. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Bruice TC. A quantum mechanics/molecular mechanics study of the catalytic mechanism and product specificity of viral histone lysine methyltransferase. Biochemistry. 2007;46:9743–9751. doi: 10.1021/bi700515q. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Bruice TC. Catalytic mechanism and product specificity of rubisco large subunit methyltransferase: QM/MM and MD investigations. Biochemistry. 2007;46:5505–5514. doi: 10.1021/bi700119p. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Bruice TC. Product specificity and mechanism of protein lysine methyltransferases: Insights from the histone lysine methyltransferase SET8. Biochemistry. 2008;47:6671–6677. doi: 10.1021/bi800244s. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Bruice TC. Enzymatic mechanism and product specificity of SET-domain protein lysine methyltransferases. Proc Natl Acad Sci USA. 2008;105:5728–5732. doi: 10.1073/pnas.0801788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo HB, Guo H. Mechanism of histone methylation catalyzed by protein lysine methyltransferase SET7/9 and origin of product specificity. Proc Natl Acad Sci USA. 2007;104:8797–8802. doi: 10.1073/pnas.0702981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dirk LM, et al. Kinetic manifestation of processivity during multiple methylations catalyzed by SET domain protein methyltransferases. Biochemistry. 2007;46:3905–3915. doi: 10.1021/bi6023644. [DOI] [PubMed] [Google Scholar]
- 30.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 31.Patnaik D, et al. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J Biol Chem. 2004;279:53248–53258. doi: 10.1074/jbc.M409604200. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Miner W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Kabsch K. Automatic indexing of rotation diffraction patterns. J Appl Crystallogr. 1998;21:67–71. [Google Scholar]
- 34.Vagin A, Teplyakov A. An approach to multicopy search in molecular replacement. Acta Crystallogr D. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases. Anal Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.